Chapter 6 Baits
This chapter introduce the baits selection in collaboration with arbor bioscience staff.
6.1 First design
I have finished the bait design and analysis, and you will receive an invitation to a Dropbox folder to access the data. Briefly, I:
- used Repeat Masker to soft-masked the 1.390 input sequences for simple repeats and those in the Clusiaceae repeat database; 2.29% masked (all simple and low complexity repeats)
- designed 80 nt baits with 2x tiling density = 29,426 raw unfiltered baits
- BLASTed each bait candidate against the two provided genomes
- kept only baits passing “Moderate” BLAST filtering, that were Repeat Masked, had GC content > 20% and < 80% = 20,719 baits
These baits cover 76.67% of desired target positions with at least 1 bait, with 91.5% within 100bp of a bait. If 719 baits are removed, it will fit into our smallest kit (1-20K); you can select the ones to remove, or I can filter them based on GC/deltaG, or remove loci with poor bait coverage. Please review the files and let me know any changes you would like to make, or if you have any questions.
Brian
More info are available in the pdf. The main objective so is to select 719 probes to be removed. We thus want to remove the 719 baits by filtering out loci with poor bait coverage at the exception of loci included in targets with candidate genes
(see figure 6.1).
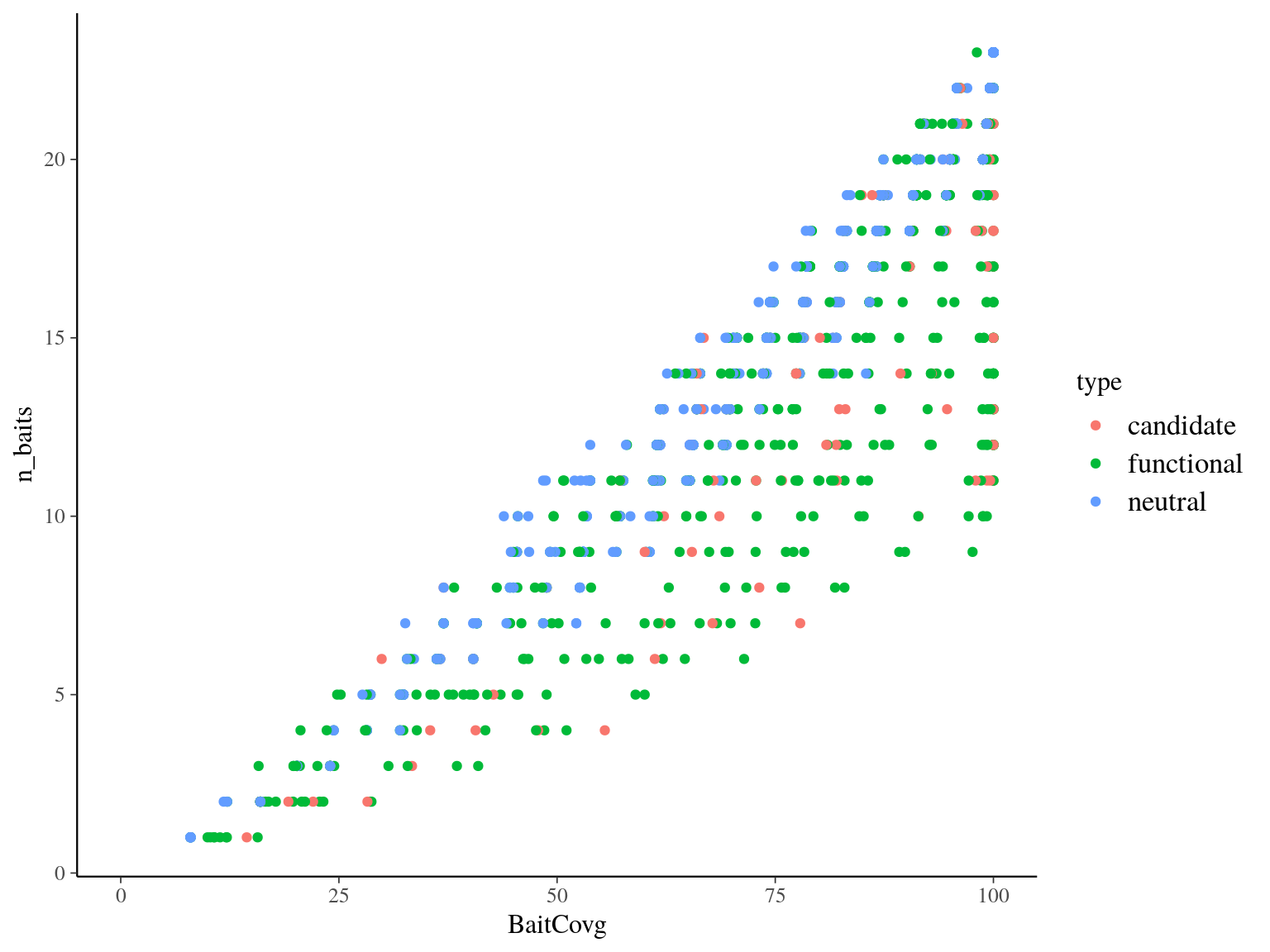
Figure 6.1: Targets baits headcount and coverage by type.