Chapter 21 Functional genomics
21.1 Genotype
We used the following model to asses genetic varaince associated to leaf functional traits once we accounted for ontogenetic variation through DBH:
\[\begin{equation} Trait_{t,p,plot,i} \sim \mathcal{logN}(log(\alpha_{t,p}.a_{t,i}.\delta_{plot}.\frac{DBH_i}{{\beta_{DBH}}_{t,p} + DBH_i}), \sigma_3) \\ a_{t,i} \sim \mathcal{MVlogN}(log(1), \sigma_2.K) \\ \delta_{plot}\sim \mathcal {logN}(log(1),\sigma_3) \tag{21.1} \end{equation}\]
We fitted the equivalent model with following priors:
\[\begin{equation} Trait_{t,p,plot,i} \sim \mathcal{logN}(log(\alpha_{t,p}.\hat{a_{t,i}}.\hat{\delta_{plot}}.\frac{DBH_i}{{\beta_{DBH}}_{t,p} + DBH_i}), \sigma_3) \\ \hat{a_{t,i}} = e^{\sigma_2.A.\epsilon_{1,i}} \\ \hat{\delta_{plot}} = e^{\sigma_3.\epsilon_{2,plot}} \\ \epsilon_{1,i} \sim \mathcal N(0,1) \\ \epsilon_{2,plot} \sim \mathcal N(0,1) \\ ~ \\ \beta_{DBH} \sim \mathcal{logN}(log(1),1) \\ (\sigma_1,\sigma_2,\sigma_3) \sim \mathcal{N}_T^3(0,1) \\ ~ \\ V_P = Var(log(\alpha_{t,p})) \\ V_G = \sigma_2^2 \\ V_{DBH} = Var(log(\frac{DBH_i}{{\beta_{DBH}}_{t,p} + DBH_i})) \\ V_{plot}= \sigma_3^2 \\ V_R = \sigma_1^2 \tag{21.2} \end{equation}\]
| Variable | Parameter | Population | Estimate | \(\sigma\) | \(\hat{R}\) |
|---|---|---|---|---|---|
| LMA | alpha | S. globulifera Paracou | 1.1807498 | 0.0578560 | 1.0009385 |
| LMA | alpha | S. globulifera Regina | 1.0461512 | 0.0958232 | 1.0003358 |
| LMA | alpha | S. sp1 | 1.3452142 | 0.0669890 | 1.0006916 |
| LMA | betaDBH | S. globulifera Paracou | 5.0648601 | 1.3139445 | 1.0011020 |
| LMA | betaDBH | S. globulifera Regina | 5.1442279 | 2.6214540 | 0.9995415 |
| LMA | betaDBH | S. sp1 | 10.2736193 | 1.5053995 | 1.0008990 |
| LMA | Vp | 0.0083969 | 0.0052189 | 1.0004351 | |
| LMA | Vg | 0.0038228 | 0.0050892 | 1.0323247 | |
| LMA | Vr | 0.0397888 | 0.0052511 | 1.0243133 | |
| LDMC | alpha | S. globulifera Paracou | 1.0371356 | 0.0232801 | 1.0015198 |
| LDMC | alpha | S. globulifera Regina | 1.0609020 | 0.0354084 | 1.0007120 |
| LDMC | alpha | S. sp1 | 1.1164856 | 0.0216282 | 1.0008250 |
| LDMC | betaDBH | S. globulifera Paracou | 2.4684682 | 0.4964418 | 1.0002995 |
| LDMC | betaDBH | S. globulifera Regina | 1.1810471 | 0.6827667 | 1.0001141 |
| LDMC | betaDBH | S. sp1 | 2.2410314 | 0.3836865 | 1.0002232 |
| LDMC | Vp | 0.0013943 | 0.0008707 | 0.9997950 | |
| LDMC | Vg | 0.0007099 | 0.0009889 | 1.0198329 | |
| LDMC | Vr | 0.0090948 | 0.0010619 | 1.0114796 | |
| LT | alpha | S. globulifera Paracou | 1.1532006 | 0.0821860 | 1.0016910 |
| LT | alpha | S. globulifera Regina | 1.0115487 | 0.1024935 | 1.0004089 |
| LT | alpha | S. sp1 | 1.1052087 | 0.0768998 | 1.0015918 |
| LT | betaDBH | S. globulifera Paracou | 3.6863479 | 0.9796721 | 0.9993911 |
| LT | betaDBH | S. globulifera Regina | 7.3115667 | 2.5529386 | 0.9997823 |
| LT | betaDBH | S. sp1 | 5.8899614 | 0.9448999 | 0.9998338 |
| LT | Vp | 0.0022700 | 0.0018253 | 1.0007100 | |
| LT | Vg | 0.0019843 | 0.0026028 | 1.0124590 | |
| LT | Vr | 0.0271003 | 0.0030027 | 1.0067090 | |
| invLA | alpha | S. globulifera Paracou | 0.6465685 | 0.0626731 | 1.0003266 |
| invLA | alpha | S. globulifera Regina | 0.9488486 | 0.0936893 | 1.0000253 |
| invLA | alpha | S. sp1 | 1.4601511 | 0.1498795 | 1.0002693 |
| invLA | betaDBH | S. globulifera Paracou | 5.2762936 | 2.6231096 | 1.0001882 |
| invLA | betaDBH | S. globulifera Regina | 1.2758801 | 1.2126296 | 1.0000454 |
| invLA | betaDBH | S. sp1 | 11.6018500 | 3.2034643 | 1.0003547 |
| invLA | Vp | 0.1401222 | 0.0436246 | 1.0002158 | |
| invLA | Vg | 0.0083183 | 0.0122263 | 1.0134787 | |
| invLA | Vr | 0.1507073 | 0.0153106 | 1.0068340 | |
| CC | alpha | S. globulifera Paracou | 1.0375996 | 0.0333635 | 1.0006473 |
| CC | alpha | S. globulifera Regina | 0.9758495 | 0.0437684 | 1.0002397 |
| CC | alpha | S. sp1 | 1.1316220 | 0.0320985 | 1.0009015 |
| CC | betaDBH | S. globulifera Paracou | 2.4335255 | 0.7115917 | 1.0000361 |
| CC | betaDBH | S. globulifera Regina | 1.3971612 | 0.8966888 | 0.9996574 |
| CC | betaDBH | S. sp1 | 2.6454552 | 0.5750465 | 0.9999197 |
| CC | Vp | 0.0030846 | 0.0017798 | 1.0004213 | |
| CC | Vg | 0.0014635 | 0.0020048 | 1.0292375 | |
| CC | Vr | 0.0181489 | 0.0021688 | 1.0212705 |
Figure 21.1: Traceplot.
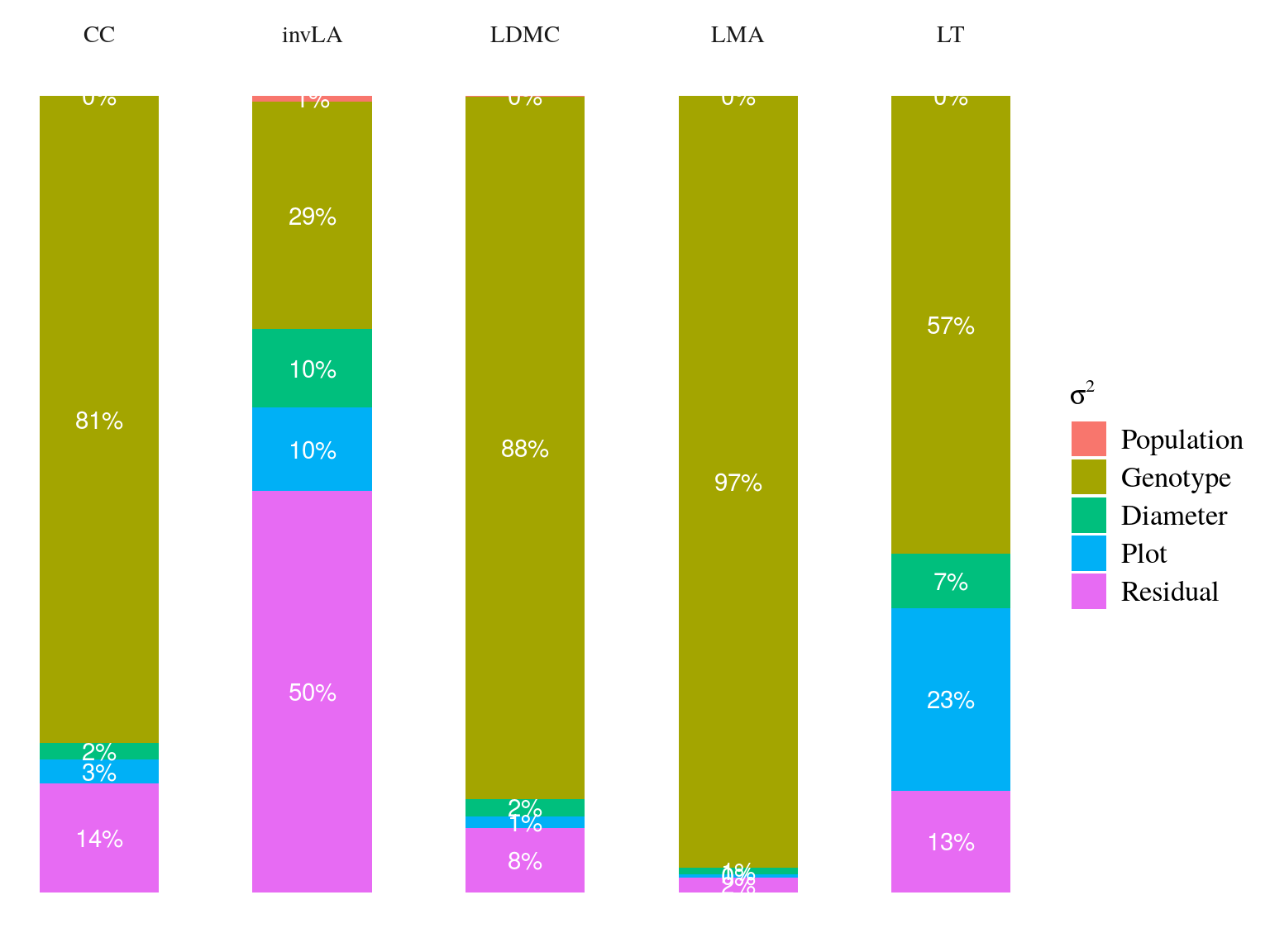
Figure 21.2: Genetic variance partitionning.