Chapter 22 Growth genomics
We sampled growth and ontogeny models developped in the chapter “Growth & Animal models”. Then, we derived growth parameters to be used in association genomics for mono- and poly-genic signals.
22.1 Gmax
We used the following growth model with a lognormal distribution to estimate individual growth potential:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \theta_{1,p,i}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\theta_{2,p}})}{\theta_{3,p}}]^2)), \sigma_1) \\ \theta_{1,p,i} \sim \mathcal {logN}(log(\theta_{1,p}), \sigma_2) \\ \theta_{2,p} \sim \mathcal {logN}(log(\theta_2),\sigma_3) \\ \theta_{3,p} \sim \mathcal {logN}(log(\theta_3),\sigma_4) \tag{22.1} \end{equation}\]
We fitted the equivalent model with following priors:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \hat{\theta_{1,p,i}}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\hat{\theta_{2,p}}})}{\hat{\theta_{3,p}}}]^2)), \sigma_1) \\ \hat{\theta_{1,p,i}} = e^{log(\theta_{1,p}) + \sigma_2.\epsilon_{1,i}} \\ \hat{\theta_{2,p}} = e^{log(\theta_2) + \sigma_3.\epsilon_{2,p}} \\ \hat{\theta_{3,p}} = e^{log(\theta_3) + \sigma_4.\epsilon_{3,p}} \\ \epsilon_{1,i} \sim \mathcal{N}(0,1) \\ \epsilon_{2,p} \sim \mathcal{N}(0,1) \\ \epsilon_{3,p} \sim \mathcal{N}(0,1) \\ ~ \\ (\theta_{1,p}, \theta_2, \theta_3) \sim \mathcal{logN}^3(log(1),1) \\ (\sigma_1, \sigma_2, \sigma_3, \sigma_4) \sim \mathcal{N}^4_T(0,1) \\ ~ \\ V_P = Var(log(\mu_p)) \\ V_R=\sigma_2^2 \tag{22.2} \end{equation}\]
| Parameter | Estimate | Standard error | \(\hat R\) | \(N_{eff}\) |
|---|---|---|---|---|
| thetap1[1] | 0.5369487 | 0.0558649 | 1.052157 | 208 |
| thetap1[2] | 0.5418062 | 0.0975853 | 1.014136 | 304 |
| thetap1[3] | 0.3671035 | 0.0243181 | 1.033224 | 229 |
| theta2 | 0.2545044 | 0.0806695 | 1.011055 | 702 |
| theta3 | 0.7014813 | 0.1053930 | 1.020937 | 491 |
| sigma[1] | 0.1335699 | 0.0941555 | 1.508783 | 22 |
| sigma[2] | 0.6703421 | 0.0406016 | 1.141318 | 62 |
| sigma[3] | 0.2739054 | 0.2875370 | 1.007774 | 972 |
| sigma[4] | 0.1336316 | 0.2239892 | 1.008755 | 1160 |
| lp__ | 362.3623923 | 267.2329411 | 1.741902 | 17 |
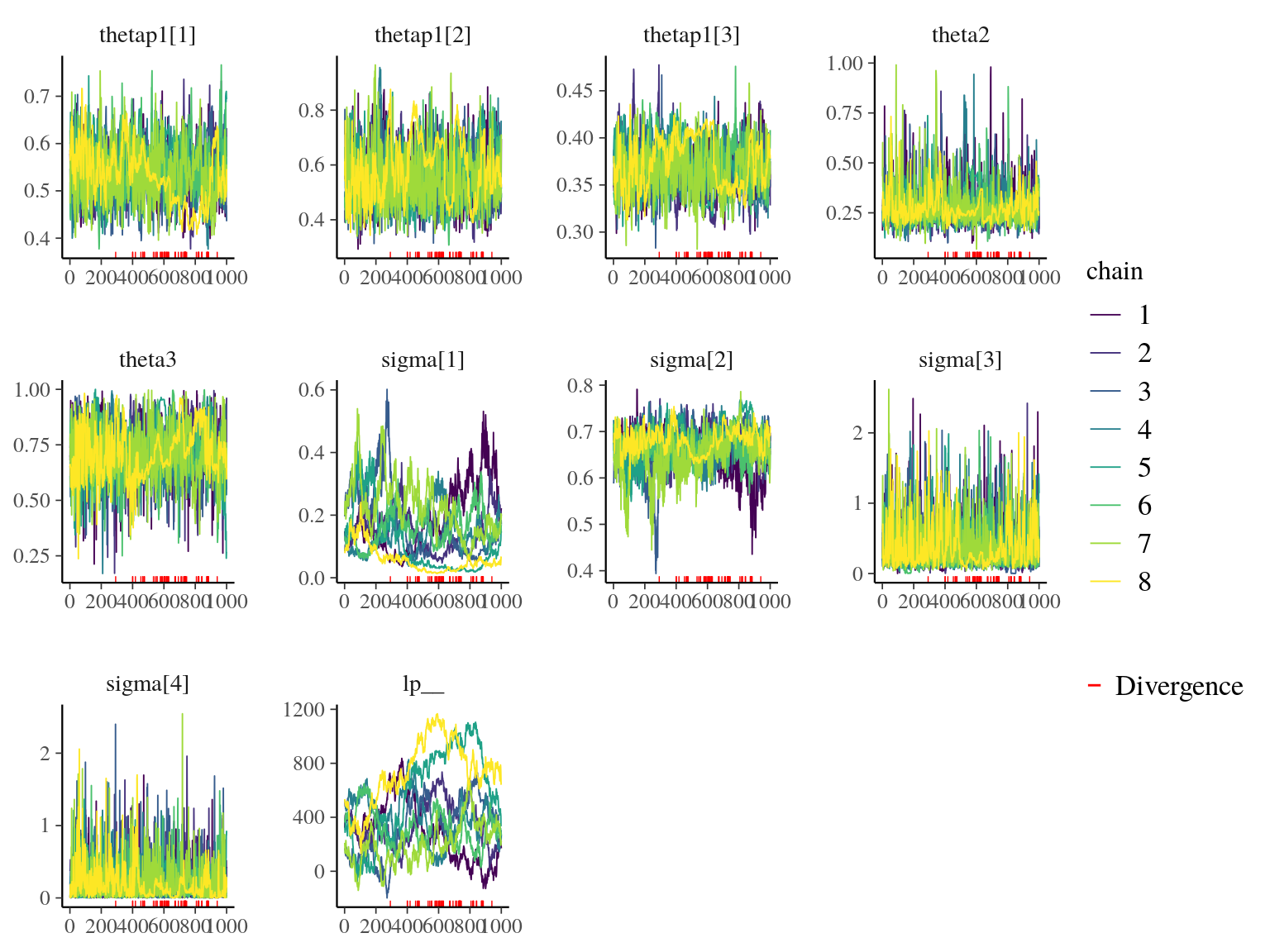
Figure 22.1: Traceplot of model parameters.
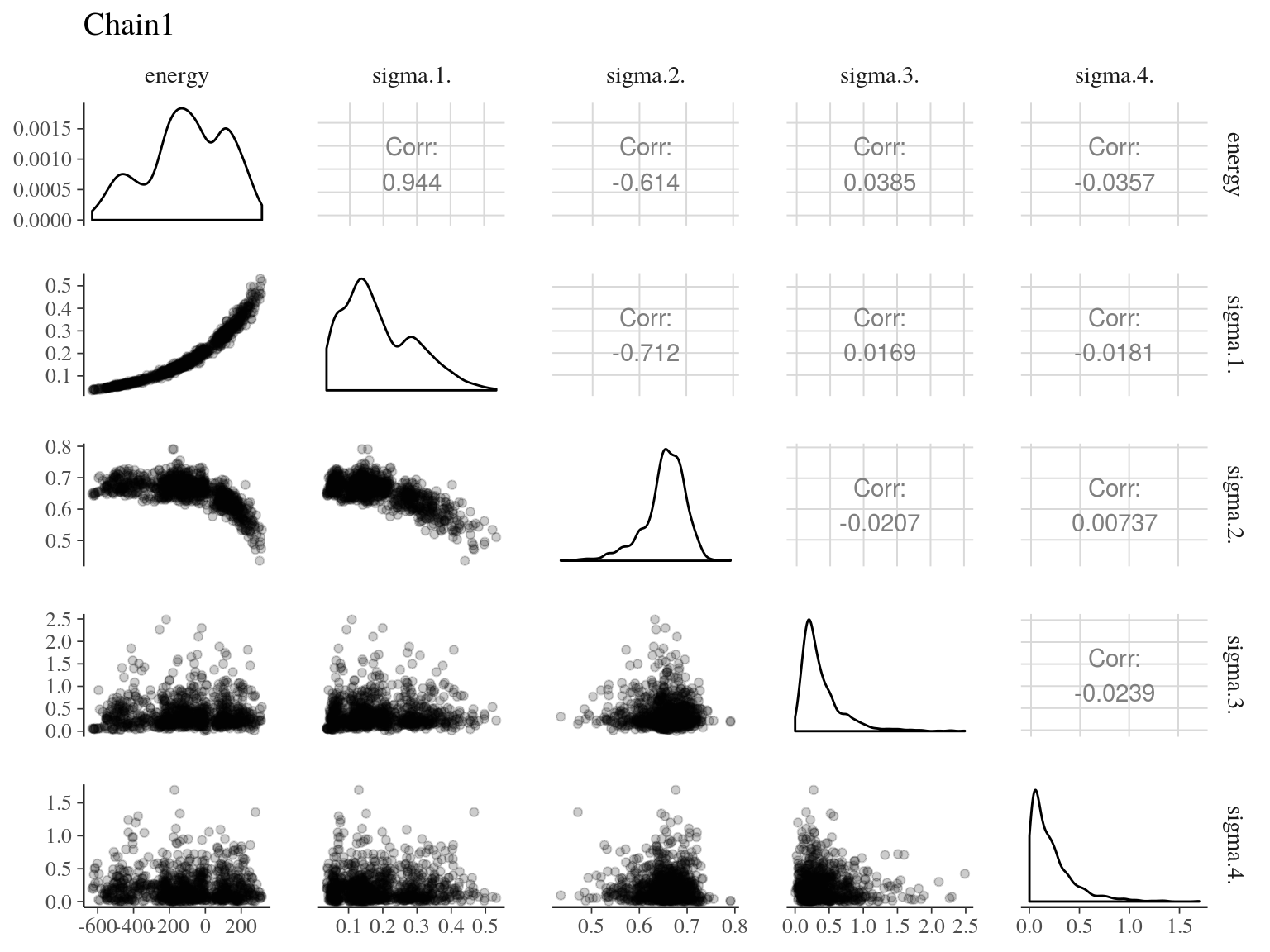
Figure 22.2: Pairs of model parameters.
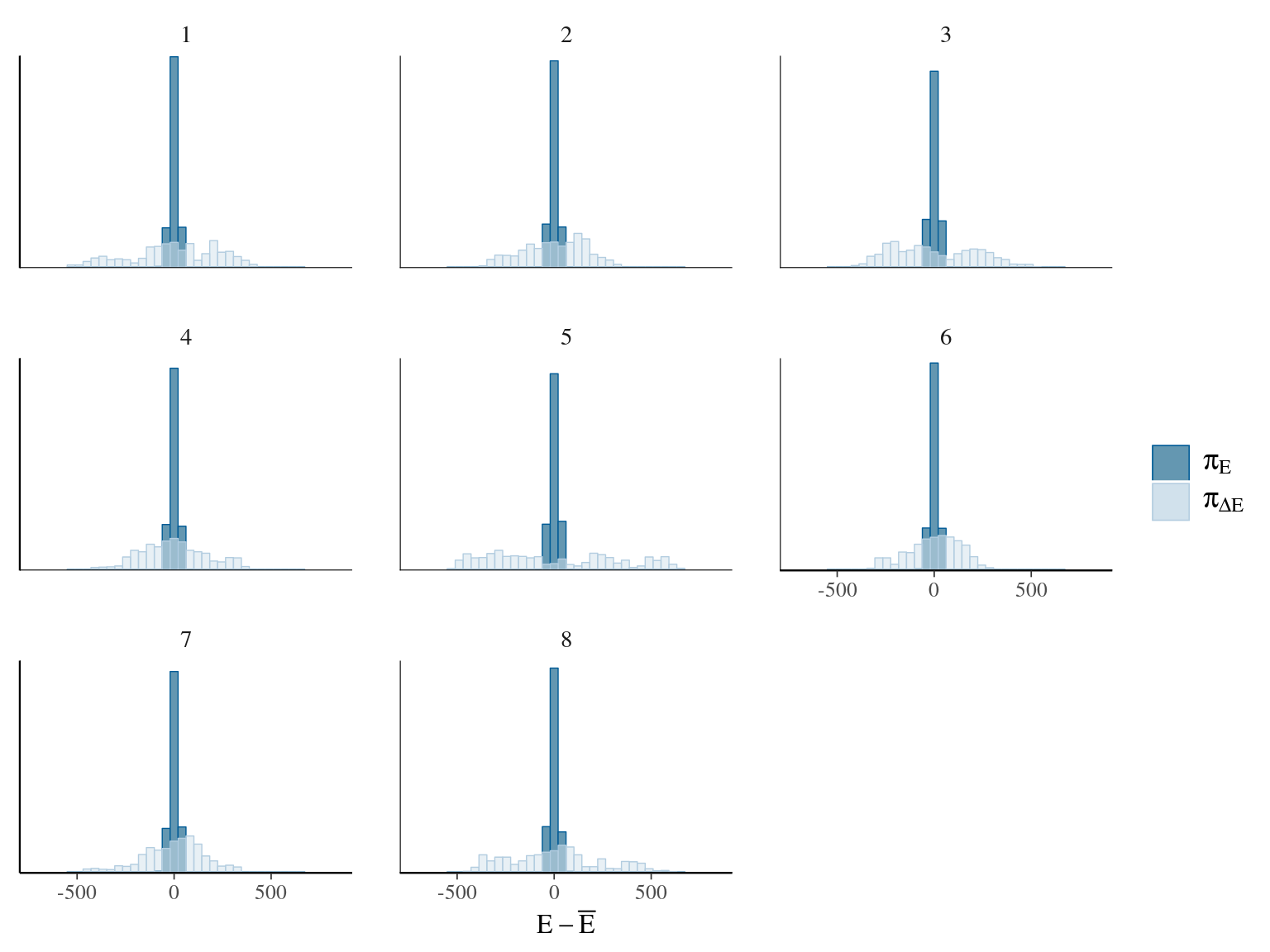
Figure 22.3: Energy of the model.
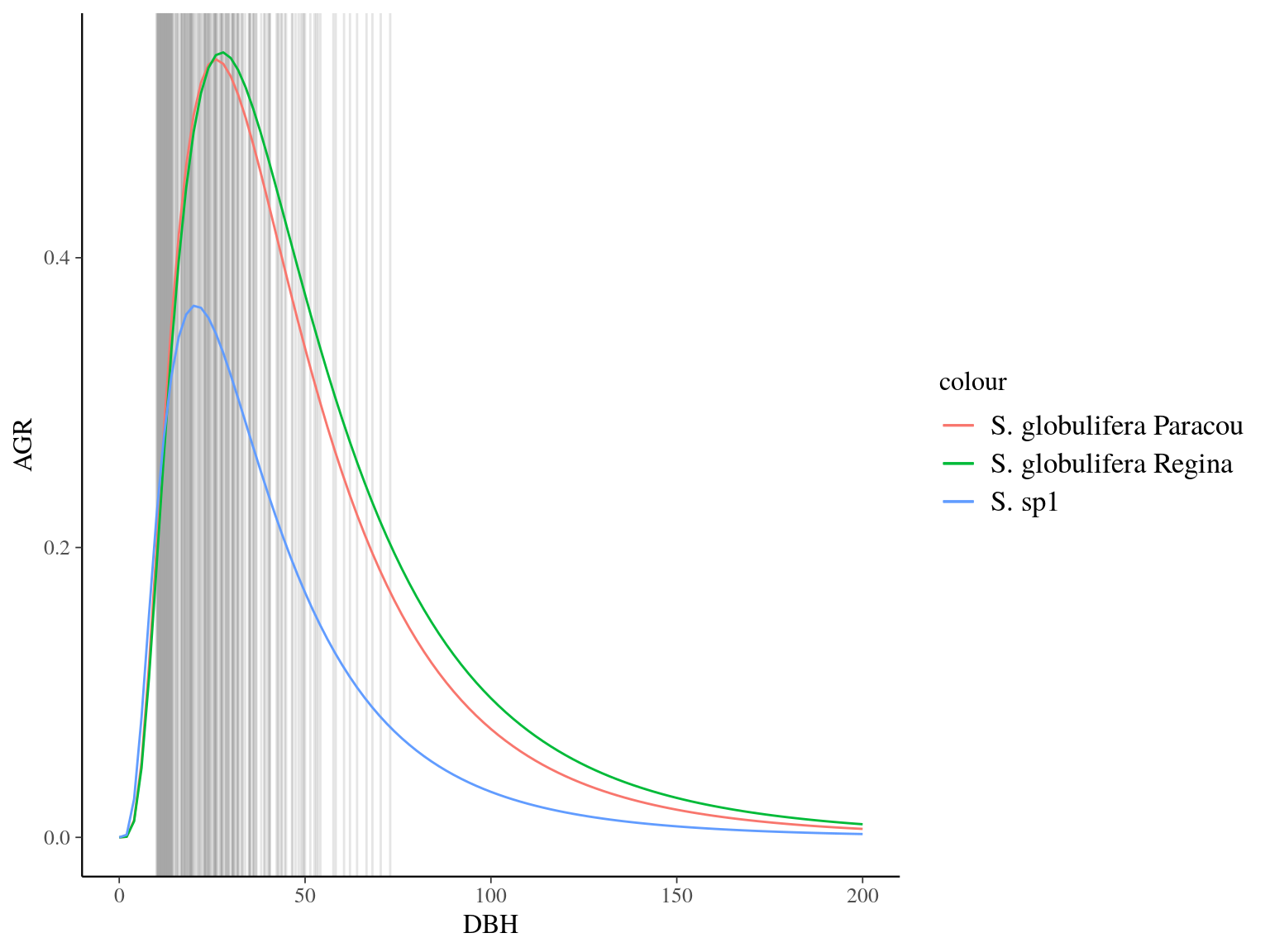
Figure 22.4: Species predicted growth curve.
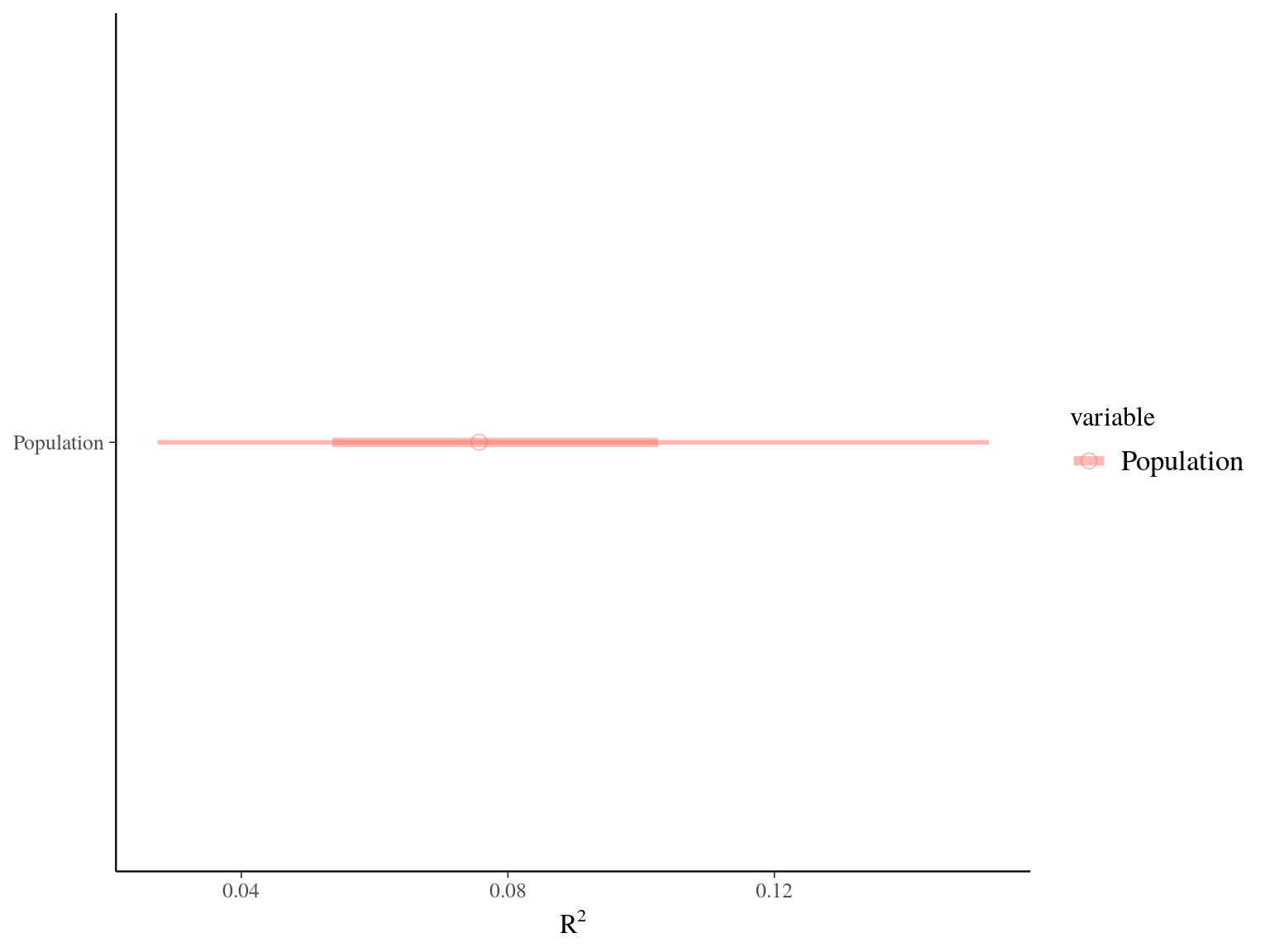
Figure 22.5: R2 for Gmax.
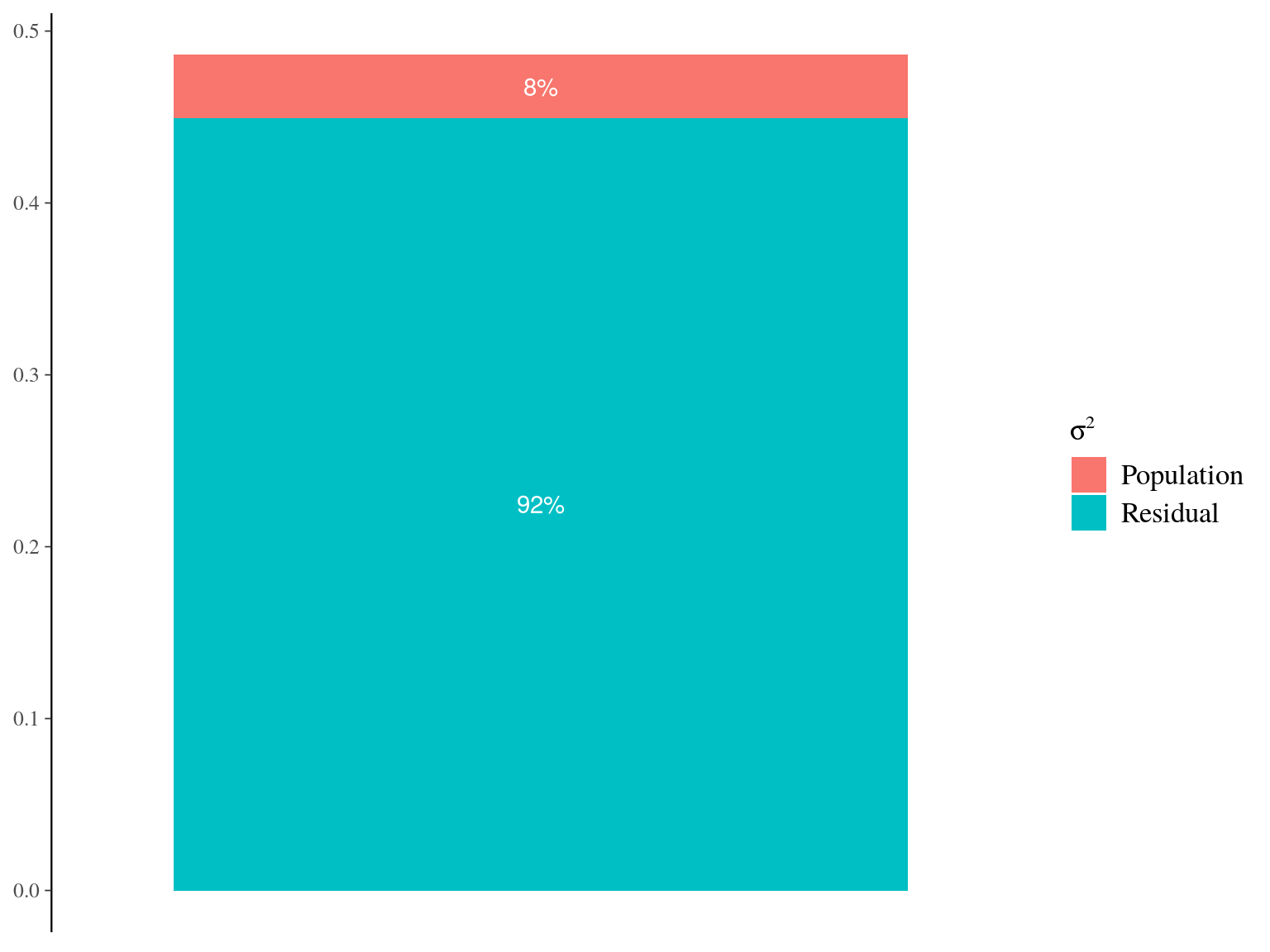
Figure 22.6: Genetic variance partitionning for Gmax.
22.2 Gmax & Genotype
We used the following growth model with a lognormal distribution to estimate individual growth potential and associated genotypic variation:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \theta_{1,p,i}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\theta_{2,p}})}{\theta_{3,p}}]^2)), \sigma_1) \\ \theta_{1,p,i} \sim \mathcal {logN}(log(\theta_{1,p}.a_{1,i}), \sigma_2) \\ \theta_{2,p} \sim \mathcal {logN}(log(\theta_2),\sigma_3) \\ \theta_{3,p} \sim \mathcal {logN}(log(\theta_3),\sigma_4) \\ a_{1,i} \sim \mathcal{MVlogN}(log(1), \sigma_5.K) \tag{22.3} \end{equation}\]
We fitted the equivalent model with following priors:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \hat{\theta_{1,p,i}}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\hat{\theta_{2,p}}})}{\hat{\theta_{3,p}}}]^2)), \sigma_1) \\ \hat{\theta_{1,p,i}} = e^{log(\theta_{1,p}.\hat{a_{1,i}}) + \sigma_2.\epsilon_{1,i}} \\ \hat{\theta_{2,p}} = e^{log(\theta_2) + \sigma_3.\epsilon_{2,p}} \\ \hat{\theta_{3,p}} = e^{log(\theta_3) + \sigma_4.\epsilon_{3,p}} \\ \hat{a_{1,i}} = e^{\sigma_5.A.\epsilon_{4,i}} \\ \epsilon_{1,i} \sim \mathcal{N}(0,1) \\ \epsilon_{2,p} \sim \mathcal{N}(0,1) \\ \epsilon_{3,p} \sim \mathcal{N}(0,1) \\ \epsilon_{4,i} \sim \mathcal{N}(0,1) \\ ~ \\ (\theta_{1,p}, \theta_2, \theta_3) \sim \mathcal{logN}^3(log(1),1) \\ (\sigma_1, \sigma_2, \sigma_3, \sigma_4, \sigma_5) \sim \mathcal{N}^5_T(0,1) \\ ~ \\ V_P = Var(log(\mu_p)) \\ V_G=\sigma_5^2\\ V_R=\sigma_2^2 \tag{22.4} \end{equation}\]
| Parameter | Estimate | Standard error | \(\hat R\) | \(N_{eff}\) |
|---|---|---|---|---|
| thetap1[1] | 0.5427844 | 0.0705194 | 1.002621 | 1588 |
| thetap1[2] | 0.5593631 | 0.1088714 | 1.002237 | 1840 |
| thetap1[3] | 0.3484395 | 0.0303648 | 1.002741 | 954 |
| theta2 | 0.2527690 | 0.0845444 | 1.005552 | 1944 |
| theta3 | 0.6963378 | 0.1047527 | 1.007877 | 1064 |
| sigma[1] | 0.1487772 | 0.0727287 | 1.256714 | 33 |
| sigma[2] | 0.4964445 | 0.1739066 | 1.098502 | 57 |
| sigma[3] | 0.2800740 | 0.2970960 | 1.000608 | 2252 |
| sigma[4] | 0.1381212 | 0.2356811 | 1.003299 | 2446 |
| sigma[5] | 0.4759487 | 0.1820254 | 1.067011 | 69 |
| lp__ | 138.6716378 | 192.6501649 | 1.379743 | 24 |
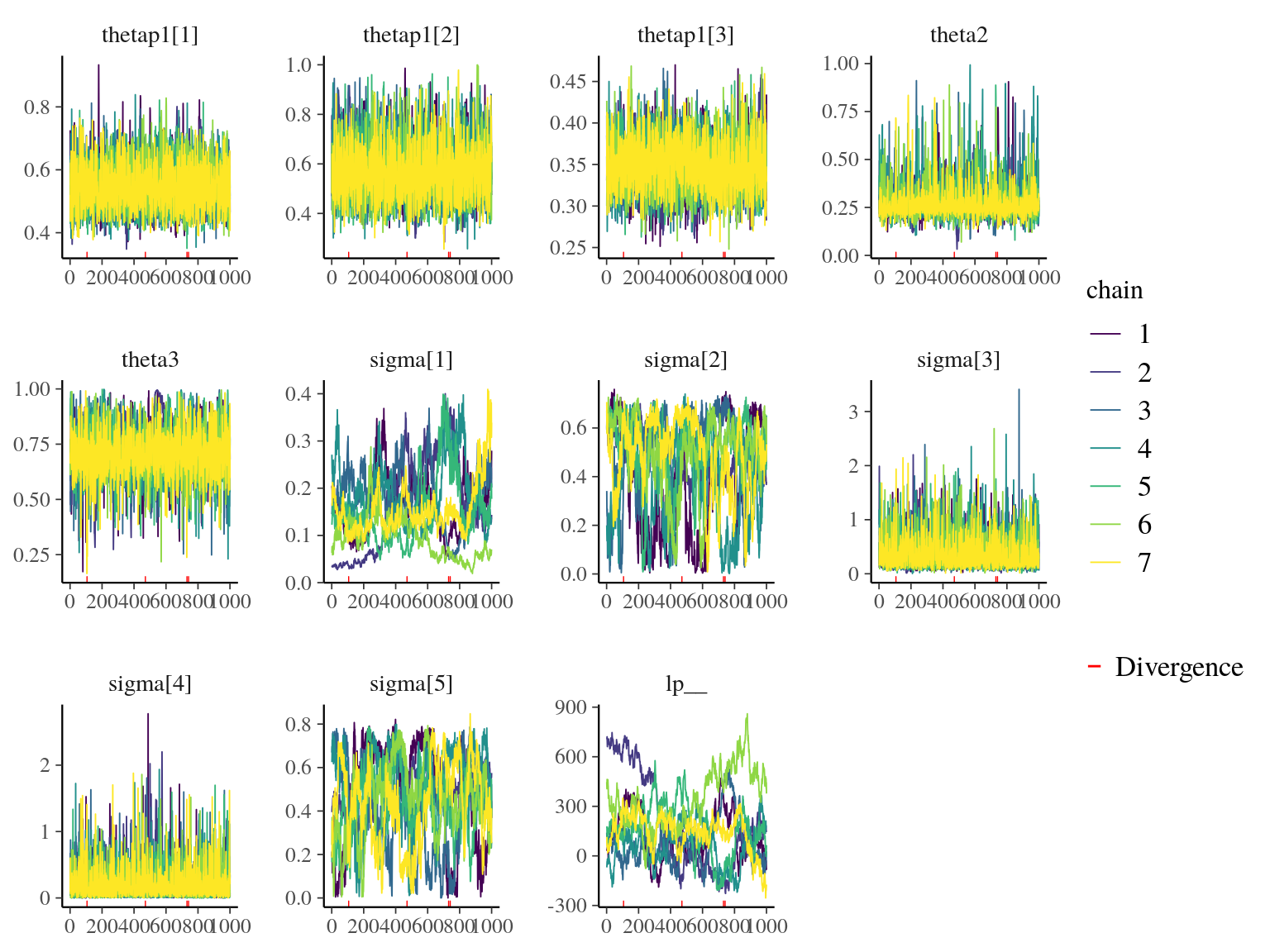
Figure 22.7: Traceplot of model parameters.
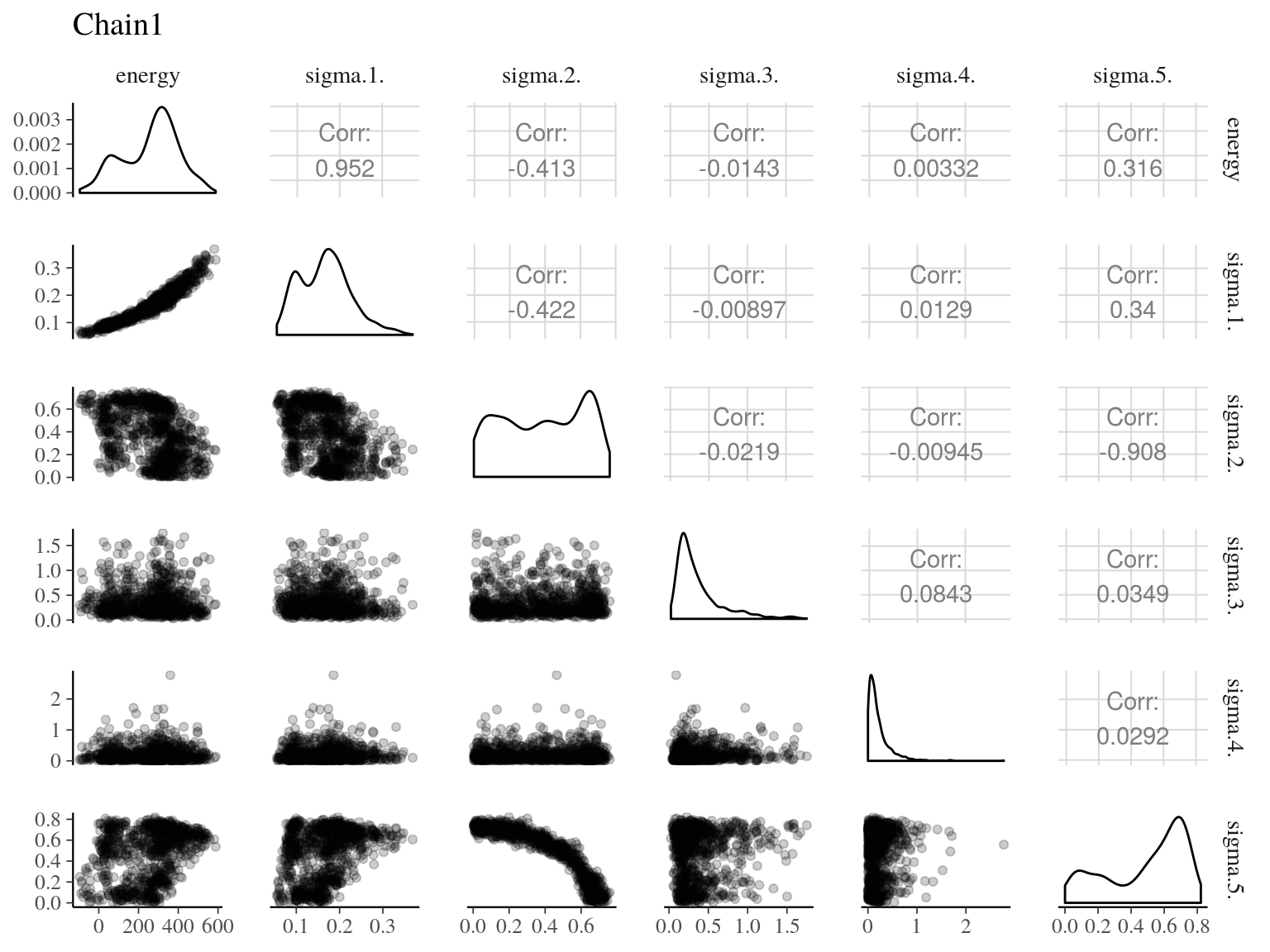
Figure 22.8: Pairs of model parameters.
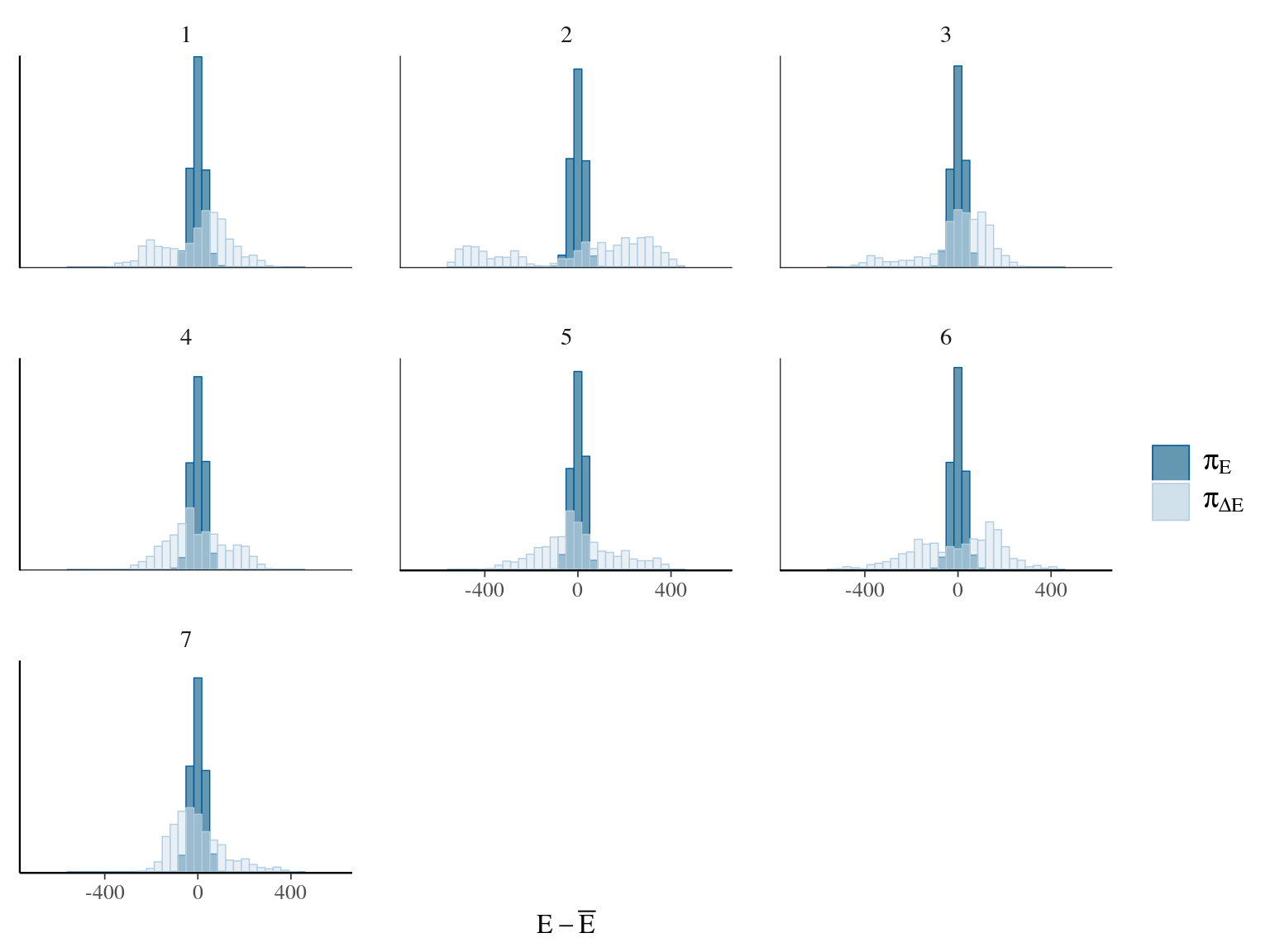
Figure 22.9: Energy of the model.
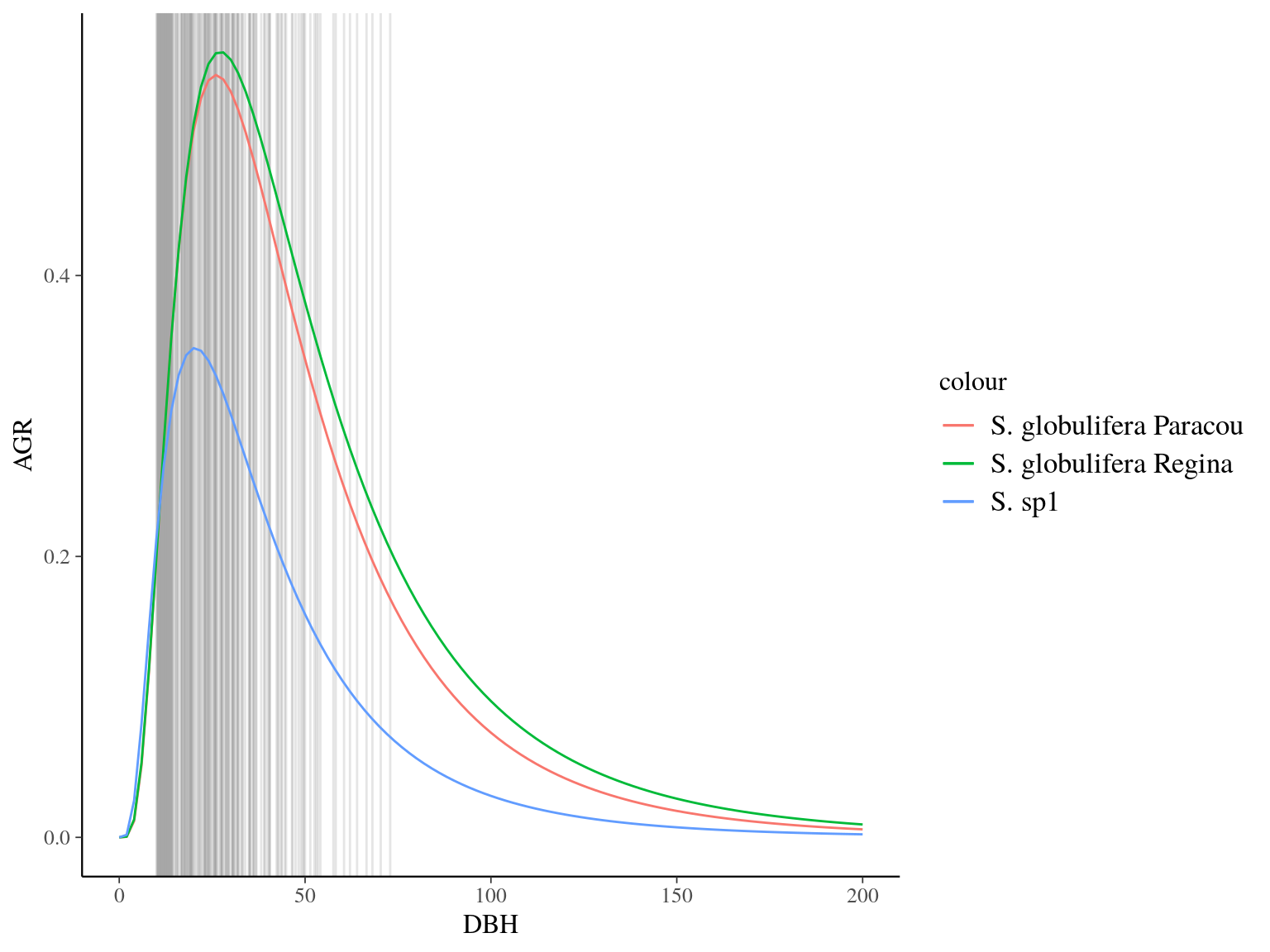
Figure 22.10: Species predicted growth curve.
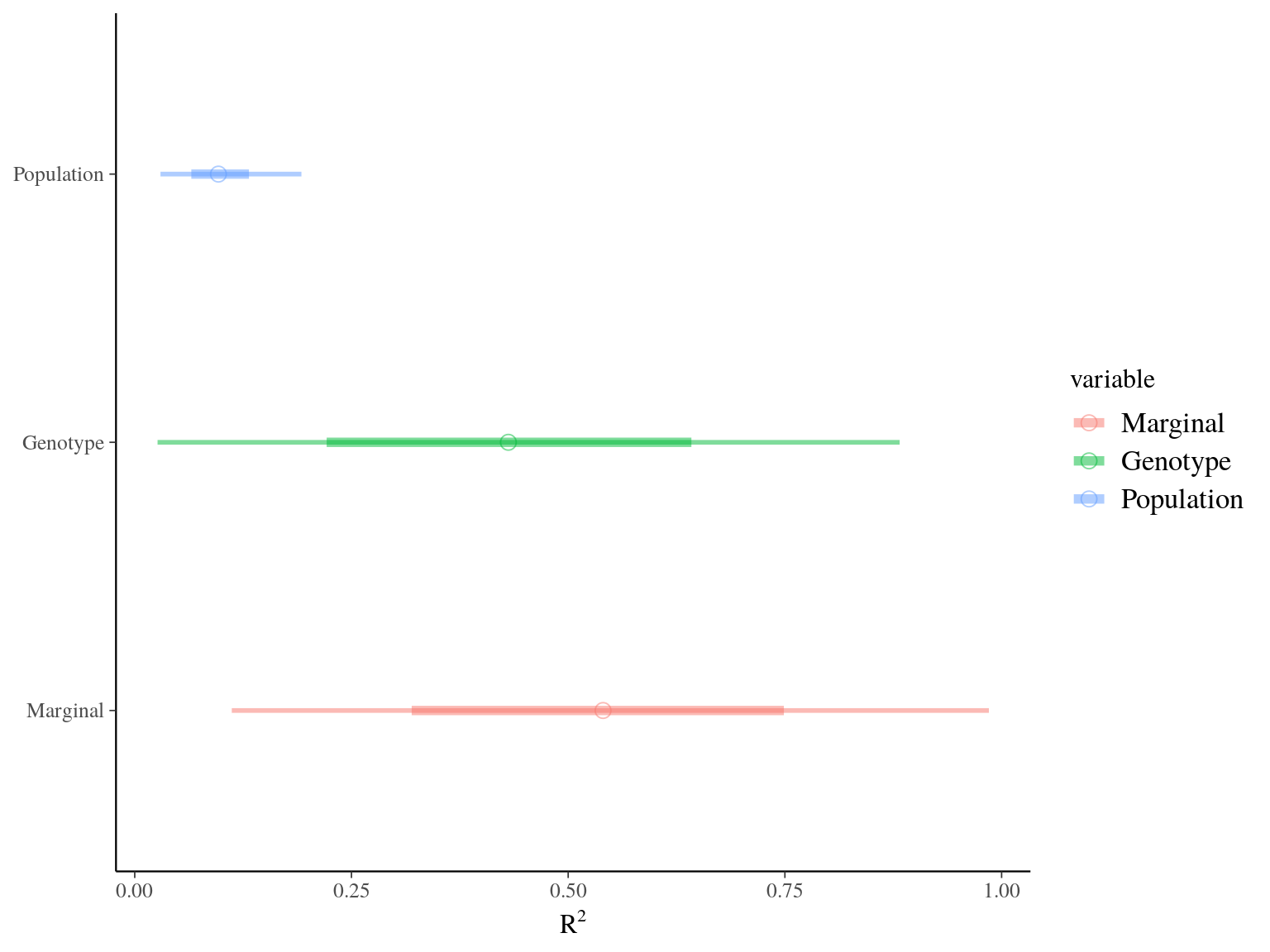
Figure 22.11: R2 for Gmax.
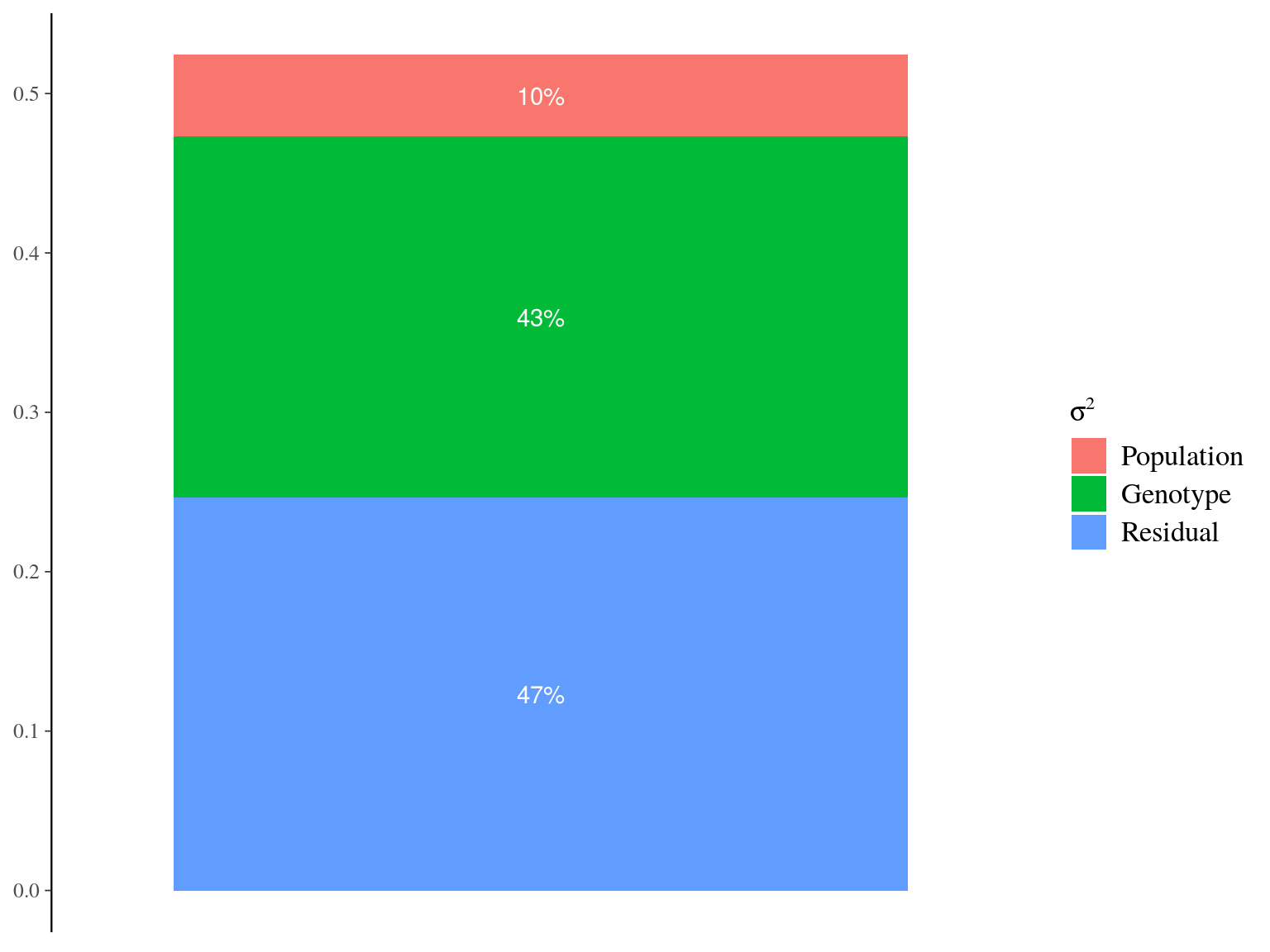
Figure 22.12: Genetic variance partitionning for Gmax.
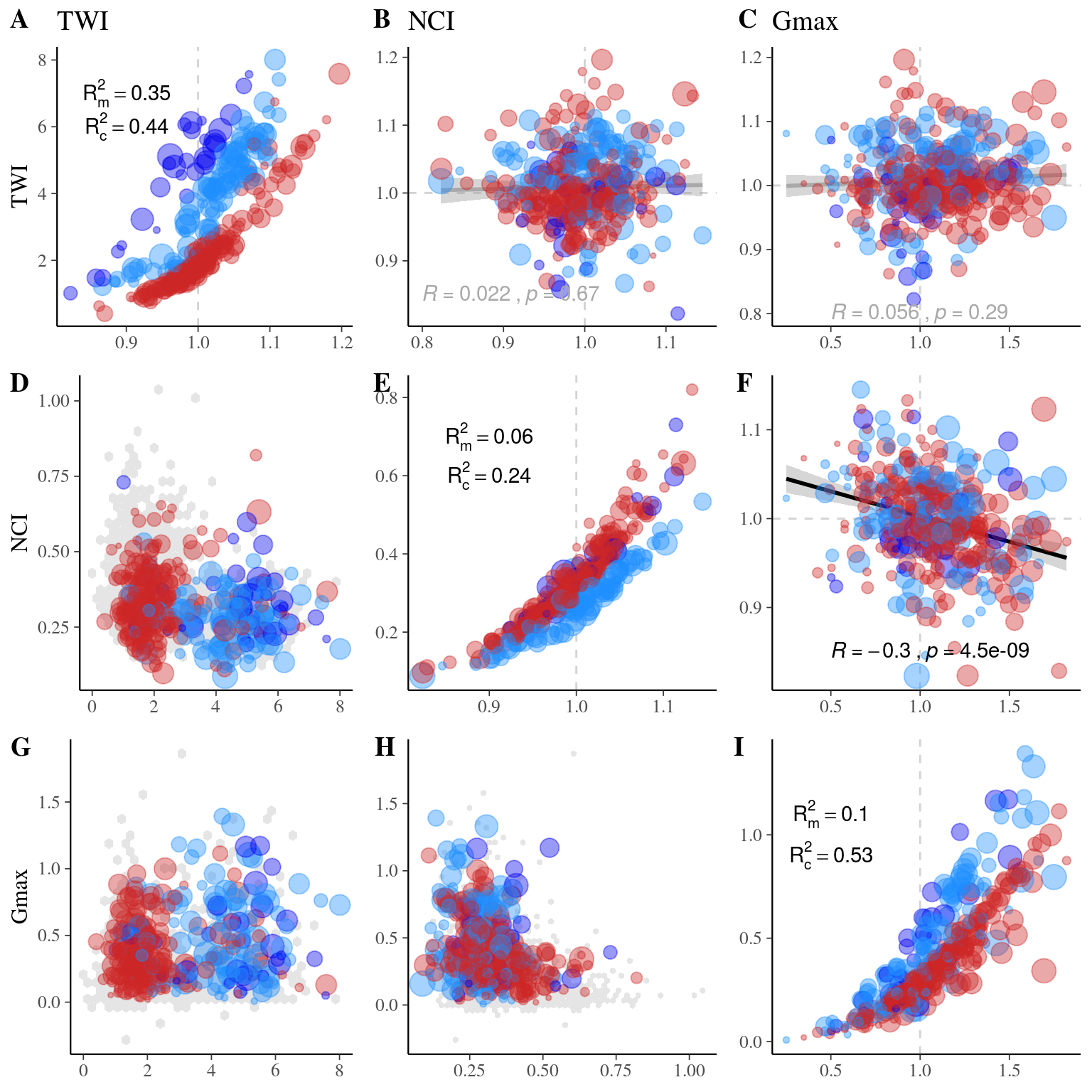
Figure 22.13: Long legend…
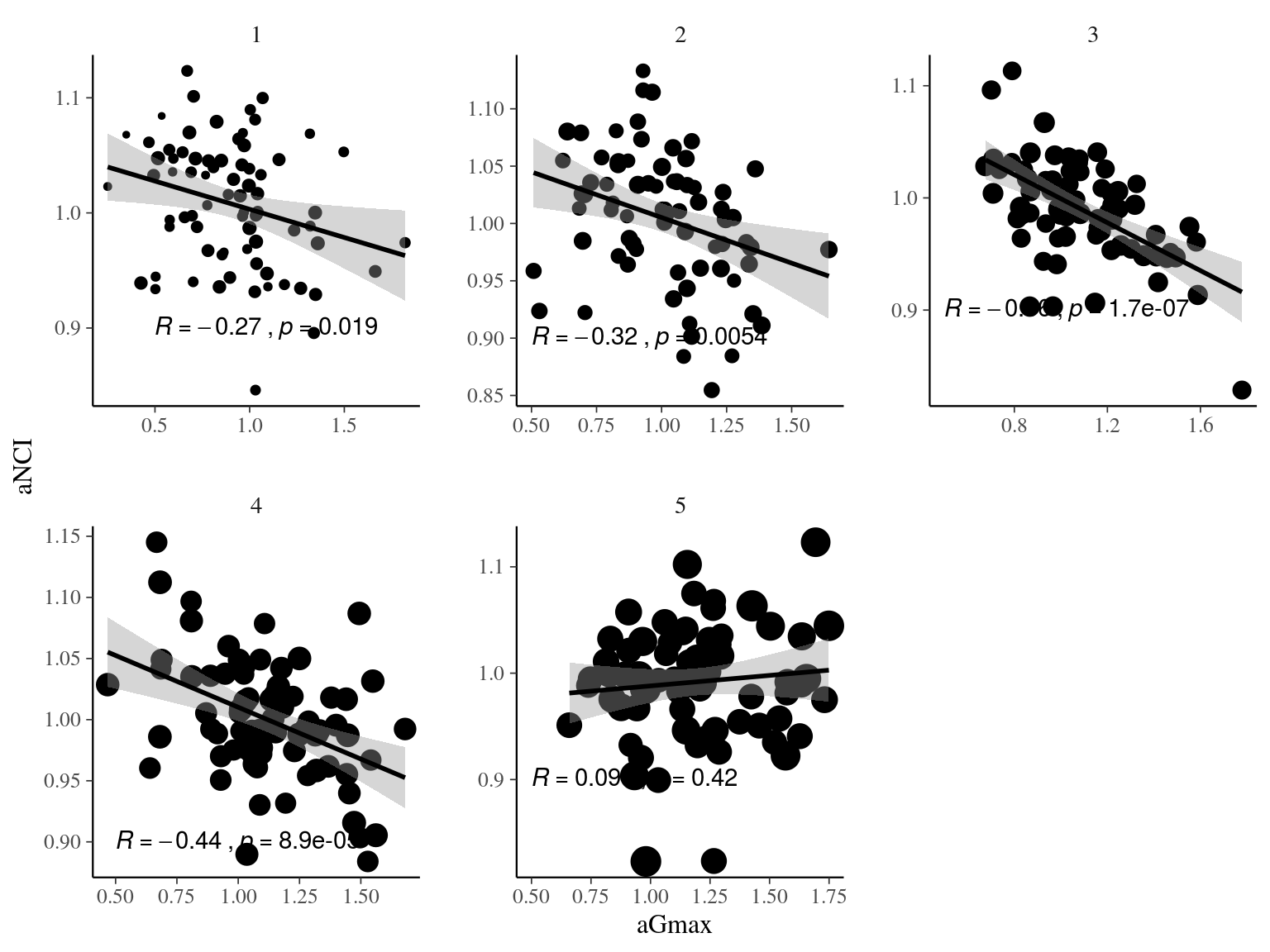
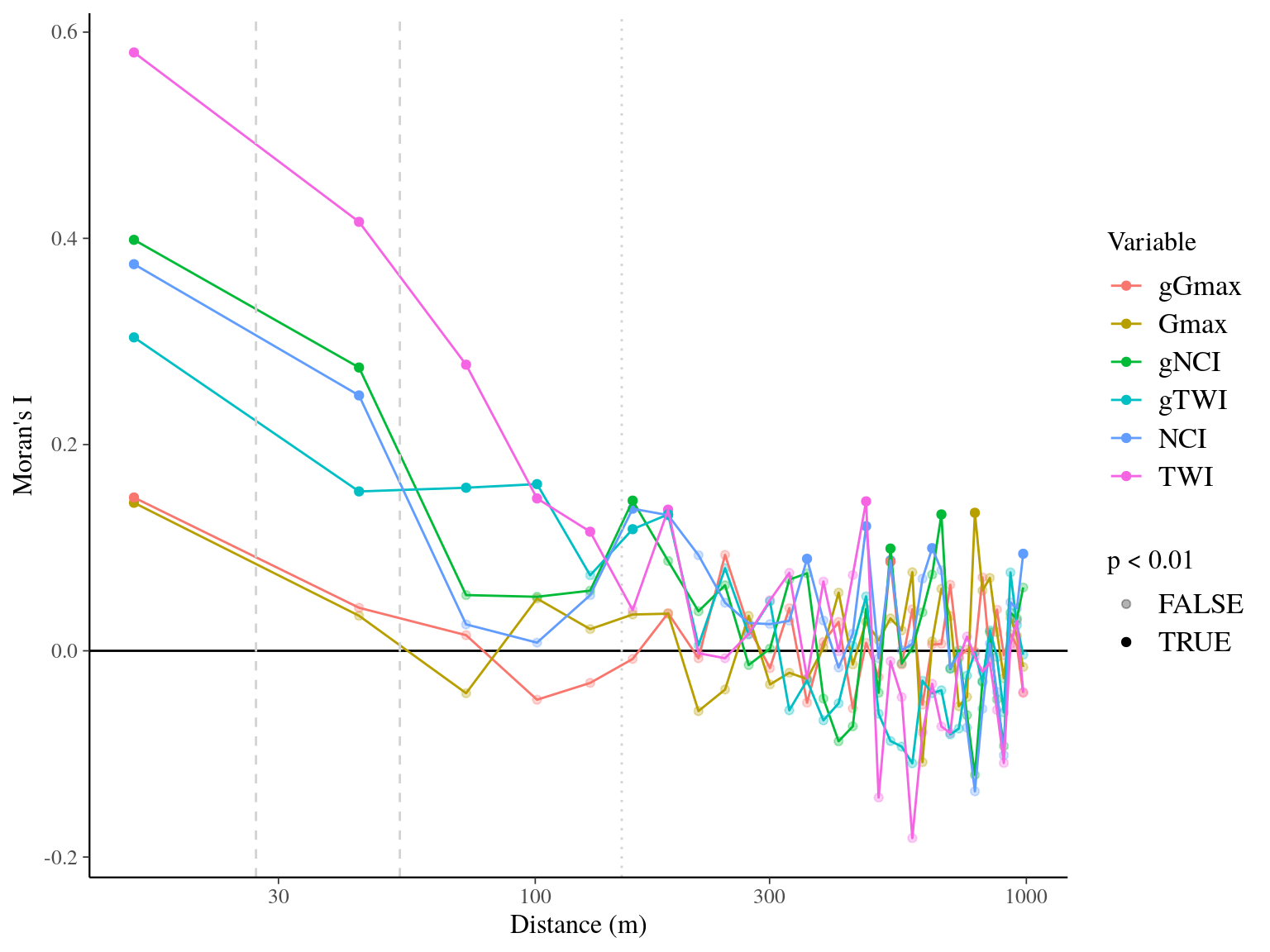
Figure 22.14: Spatial autocorrelogram (Moran’s I) of variables and associated genetic multiplicative values.
22.3 Gmax, Genotype & Environment
We used the following growth model with a lognormal distribution to estimate individual growth potential and associated genotypic variation:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \theta_{1,p,i}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\theta_{2,p}})}{\theta_{3,p}}]^2)), \sigma_1) \\ \theta_{1,p,i} \sim \mathcal {logN}(log(\theta_{1,p}.a_{1,i}. \beta_1.TWI_i.\beta_2.NCI_i), \sigma_2) \\ \theta_{2,p} \sim \mathcal {logN}(log(\theta_2),\sigma_3) \\ \theta_{3,p} \sim \mathcal {logN}(log(\theta_3),\sigma_4) \\ a_{1,i} \sim \mathcal{MVlogN}(log(1), \sigma_5.K) \tag{22.5} \end{equation}\]
We fitted the equivalent model with following priors:
\[\begin{equation} DBH_{y=today,p,i} - DBH_{y=y0,p,i} \sim \\ \mathcal{logN} (log(\sum _{y=y_0} ^{y=today} \hat{\theta_{1,p,i}}.exp(-\frac12.[\frac{log(\frac{DBH_{y,p,i}}{100.\hat{\theta_{2,p}}})}{\hat{\theta_{3,p}}}]^2)), \sigma_1) \\ \hat{\theta_{1,p,i}} = e^{log(\theta_{1,p}.\hat{a_{1,i}}. \beta_1.TWI_i.\beta_2.NCI_i) + \sigma_2.\epsilon_{1,i}} \\ \hat{\theta_{2,p}} = e^{log(\theta_2) + \sigma_3.\epsilon_{2,p}} \\ \hat{\theta_{3,p}} = e^{log(\theta_3) + \sigma_4.\epsilon_{3,p}} \\ \hat{a_{1,i}} = e^{\sigma_5.A.\epsilon_{4,i}} \\ \epsilon_{1,i} \sim \mathcal{N}(0,1) \\ \epsilon_{2,p} \sim \mathcal{N}(0,1) \\ \epsilon_{3,p} \sim \mathcal{N}(0,1) \\ \epsilon_{4,i} \sim \mathcal{N}(0,1) \\ ~ \\ (\theta_{1,p}, \theta_2, \theta_3) \sim \mathcal{logN}^3(log(1),1) \\ (\sigma_1, \sigma_2, \sigma_3, \sigma_4, \sigma_5) \sim \mathcal{N}^5_T(0,1) \\ ~ \\ V_P = Var(log(\mu_p)) \\ V_G=\sigma_5^2\\ V_{TWI} = Var(log(\beta_1.TWI_i)) \\ V_{NCI} = Var(log(\beta_2.NCI_i)) \\ V_R=\sigma_2^2 \tag{22.6} \end{equation}\]
| Parameter | Estimate | Standard error | \(\hat R\) | \(N_{eff}\) |
|---|---|---|---|---|
| thetap1[1] | 0.6819703 | 0.1750196 | 1.0017710 | 3771 |
| thetap1[2] | 0.4693647 | 0.1630881 | 1.0003561 | 3238 |
| thetap1[3] | 0.7133448 | 0.1741461 | 1.0009126 | 3999 |
| beta[1] | 1.0338461 | 0.4773498 | 1.0009493 | 5751 |
| beta[2] | 1.0231845 | 0.4821474 | 0.9998852 | 6093 |
| theta2 | 0.2212118 | 0.0872344 | 1.0031382 | 1564 |
| theta3 | 0.6542952 | 0.1080025 | 1.0011451 | 2755 |
| sigma[1] | 0.1302832 | 0.0953805 | 1.3708842 | 22 |
| sigma[2] | 0.8841277 | 0.1185958 | 1.1145255 | 81 |
| sigma[3] | 0.3078621 | 0.3262250 | 1.0020929 | 2855 |
| sigma[4] | 0.1441136 | 0.2493211 | 1.0005756 | 4162 |
| sigma[5] | 0.4157660 | 0.2270843 | 1.1430011 | 55 |
| lp__ | 184.4453908 | 241.4955933 | 1.6103979 | 13 |
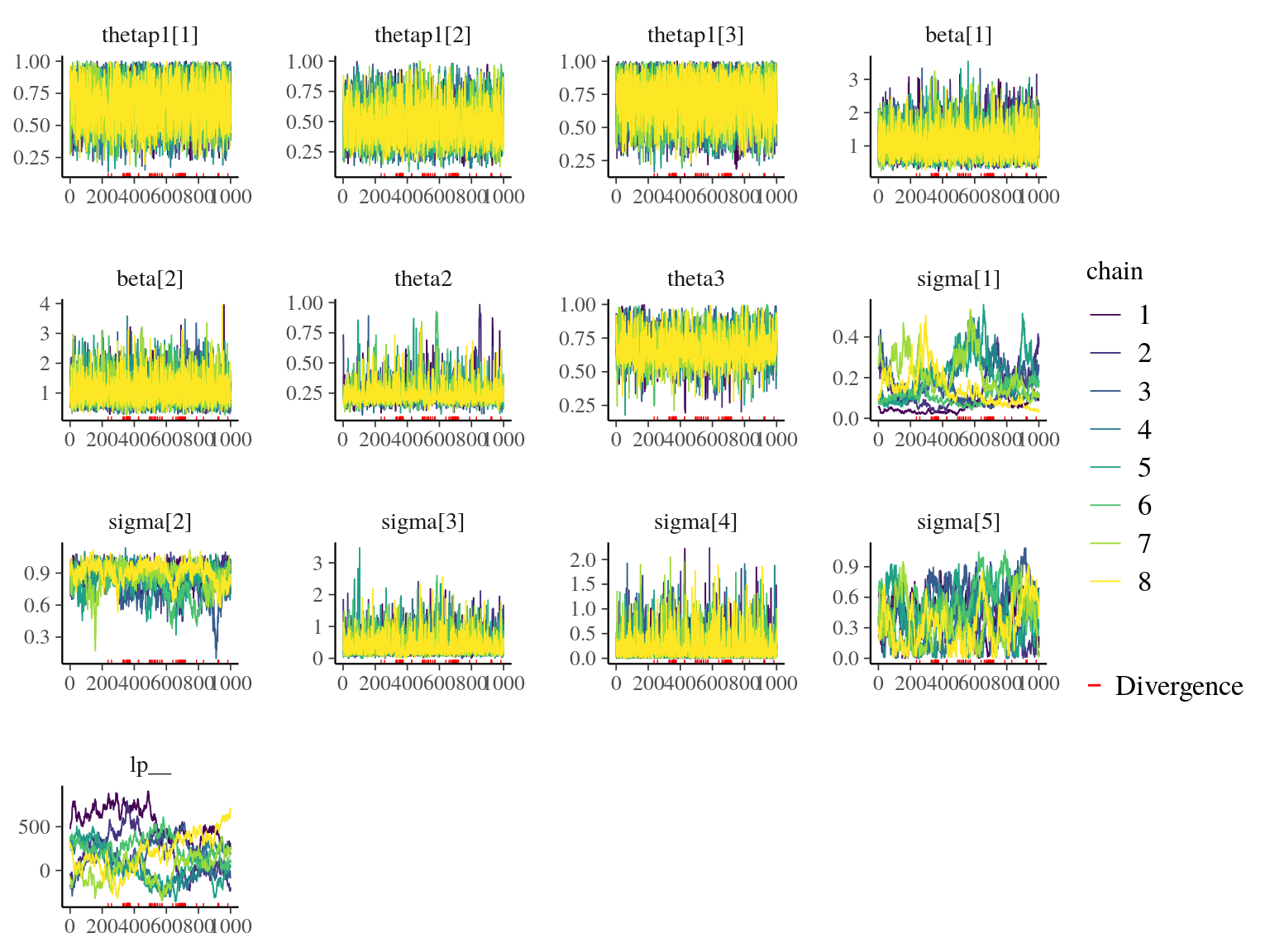
Figure 22.15: Traceplot of model parameters.
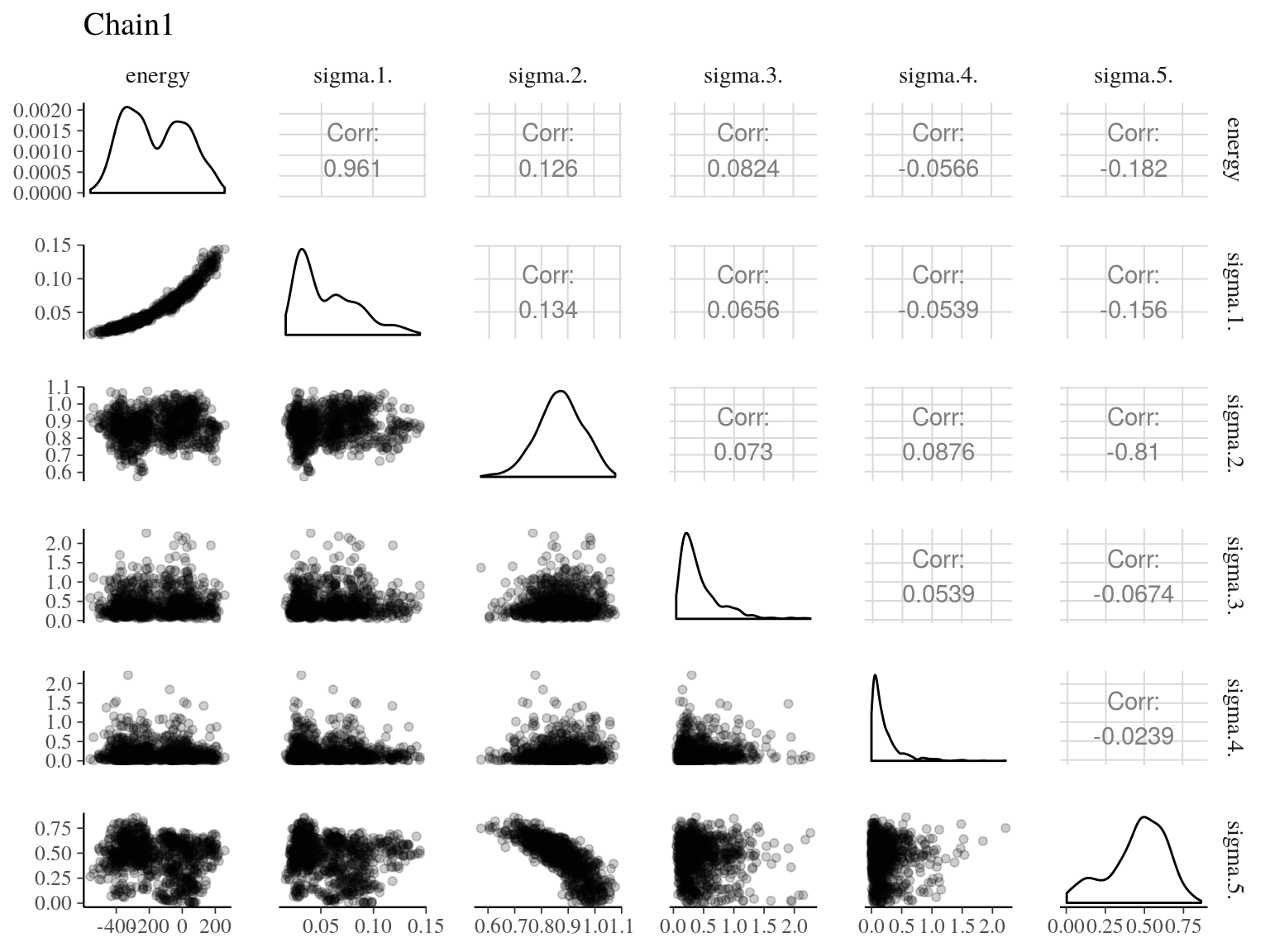
Figure 22.16: Pairs of model parameters.
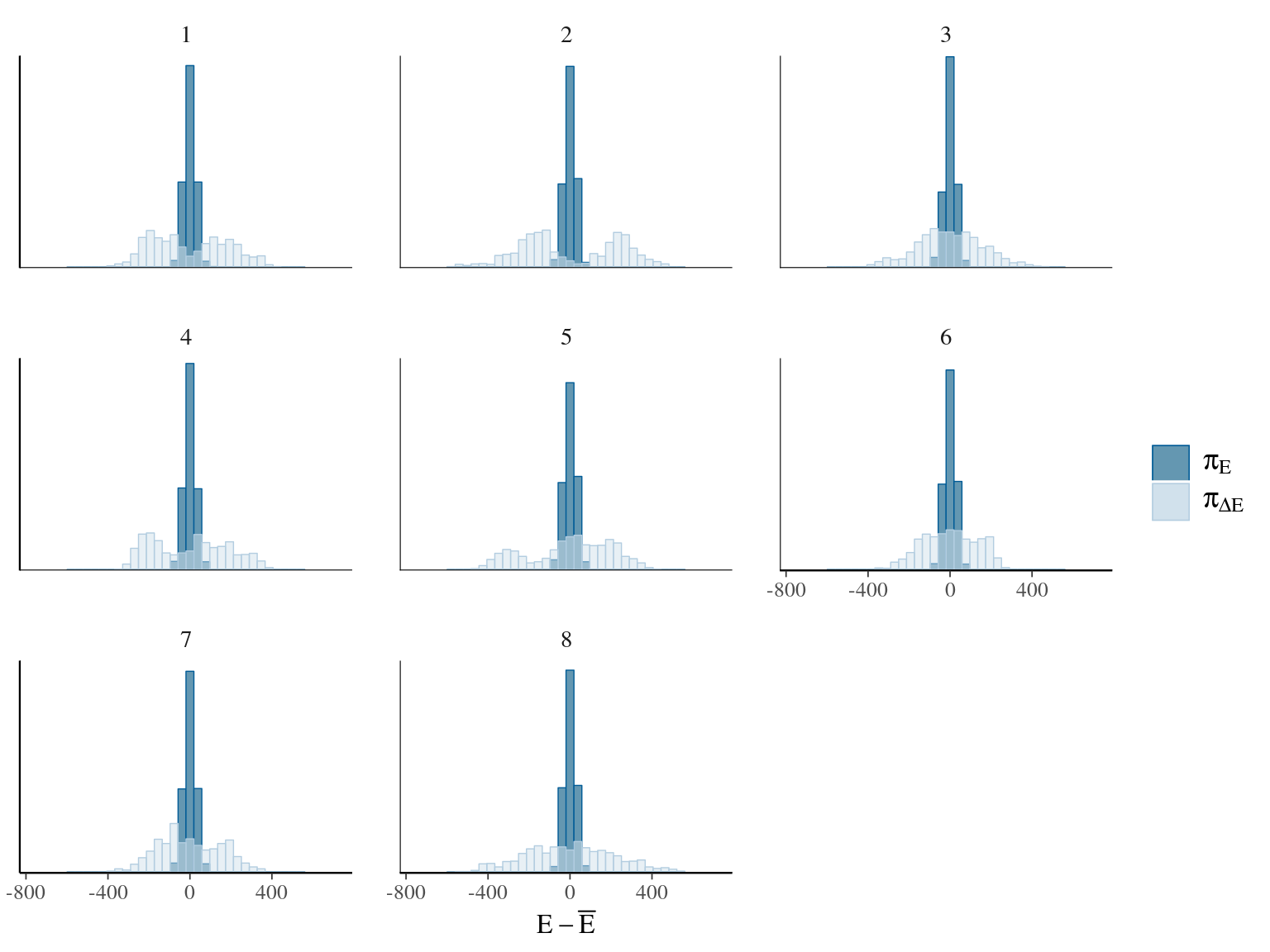
Figure 22.17: Energy of the model.
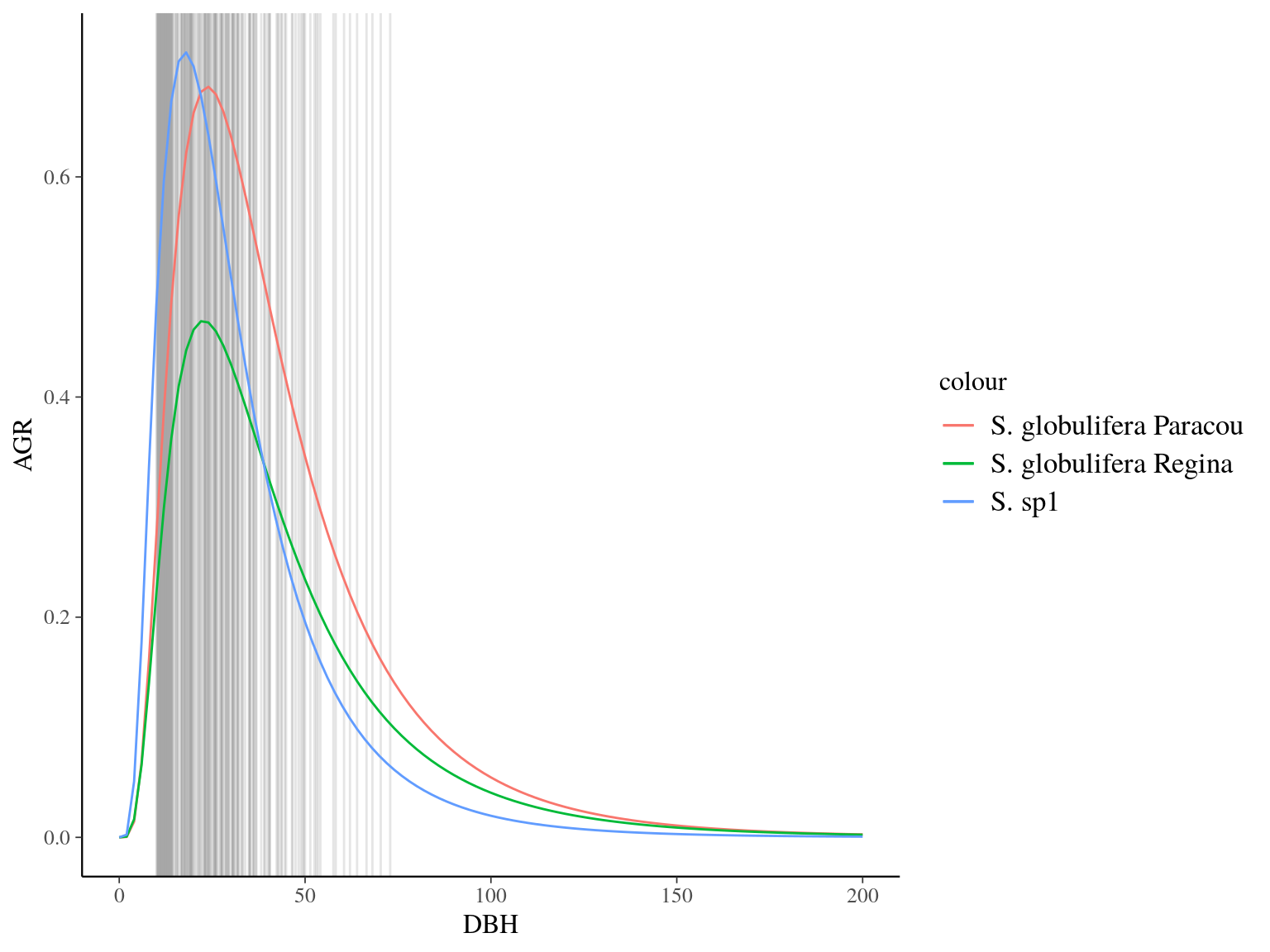
Figure 22.18: Species predicted growth curve.
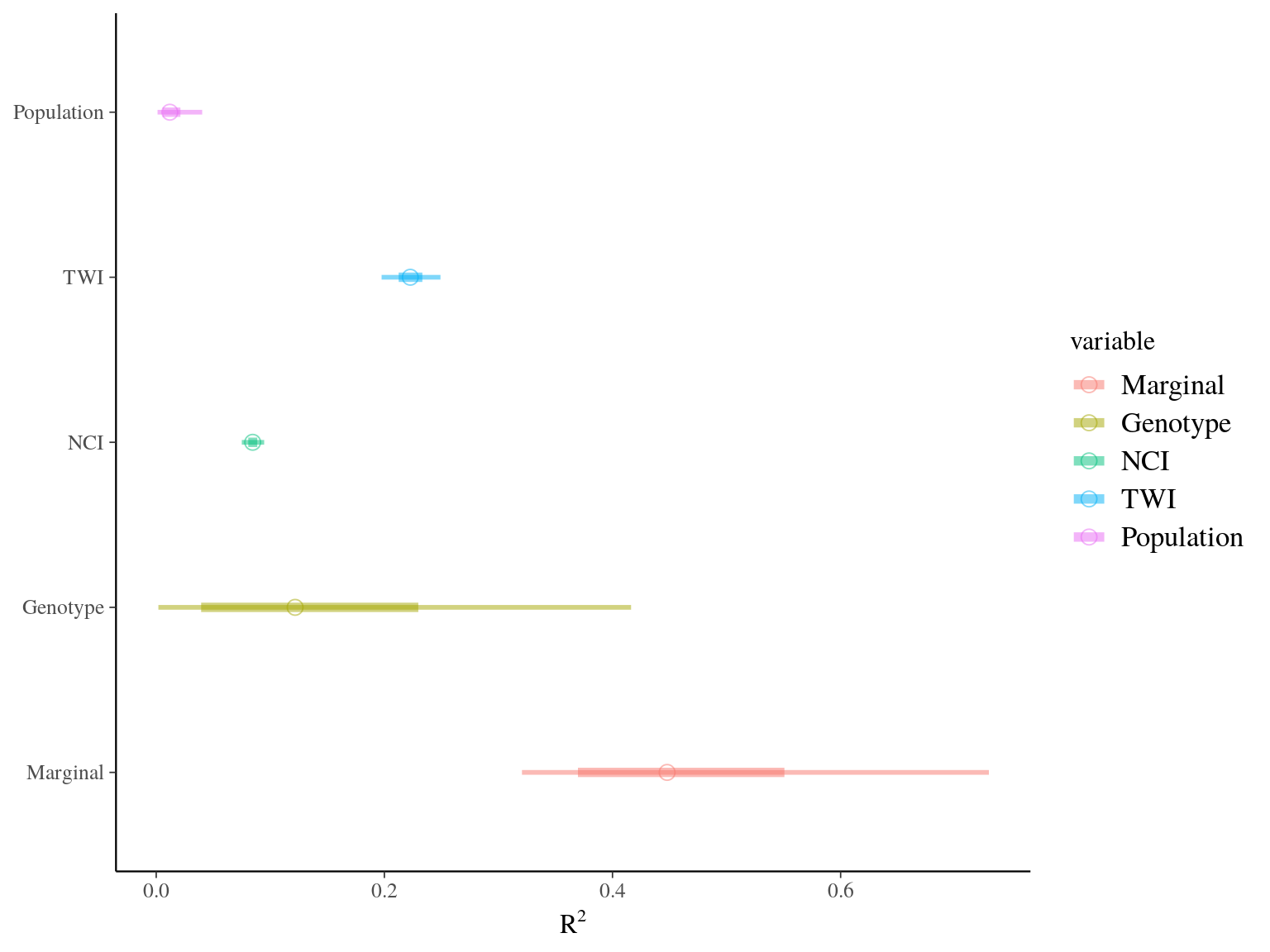
Figure 22.19: R2 for Gmax.
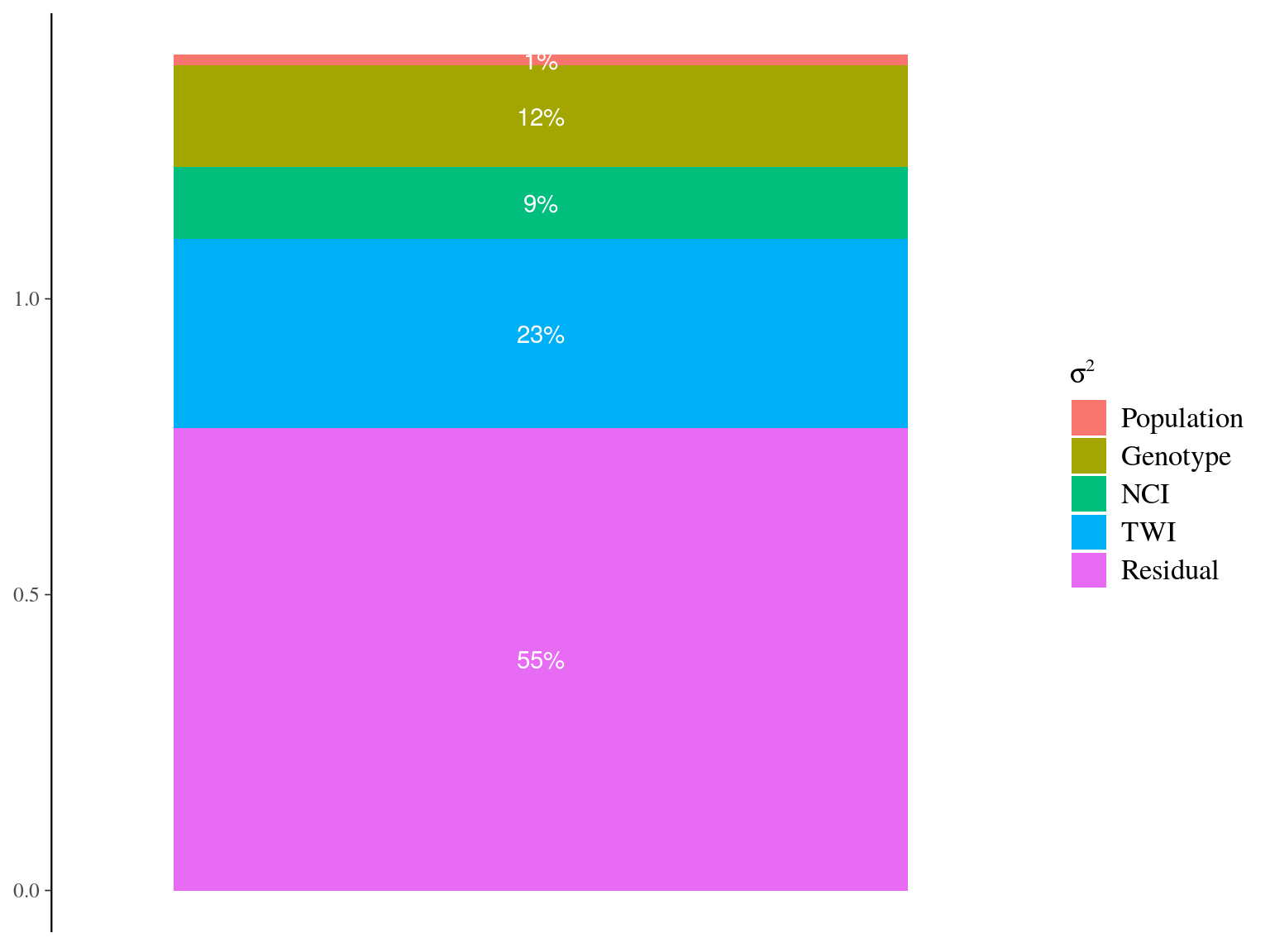
Figure 22.20: Genetic variance partitionning for ontogenetic parameters.
22.4 Associations
SNP association to growth potential \(Gmax\) has been assessed per individual with mono- and poly-genic modelling (respectivelly LMM and BSLMM, see methods in environmental genomics) for every populations.
module load bioinfo/plink-v1.90b5.3
module load bioinfo/gemma-v0.97
pops=(globuliferaTypeParacou globuliferaTypeRegina sp1)
for pop in "${pops[@]}" ;
do
plink \
--bfile growth \
--allow-extra-chr \
--keep $pop.fam \
--extract growth.prune.in \
--maf 0.05 \
--pheno gmax.phenotype \
--out $pop --make-bed ;
gemma -bfile $pop -gk 1 -o kinship.$pop ;
gemma -bfile $pop -lmm 1 \
-k output/kinship.$pop.cXX.txt \
-o LMM.$pop ;
done
cat LMM.snps | cut -f1 > snps.list
plink --bfile growth --allow-extra-chr --extract snps.list --out outliers --recode A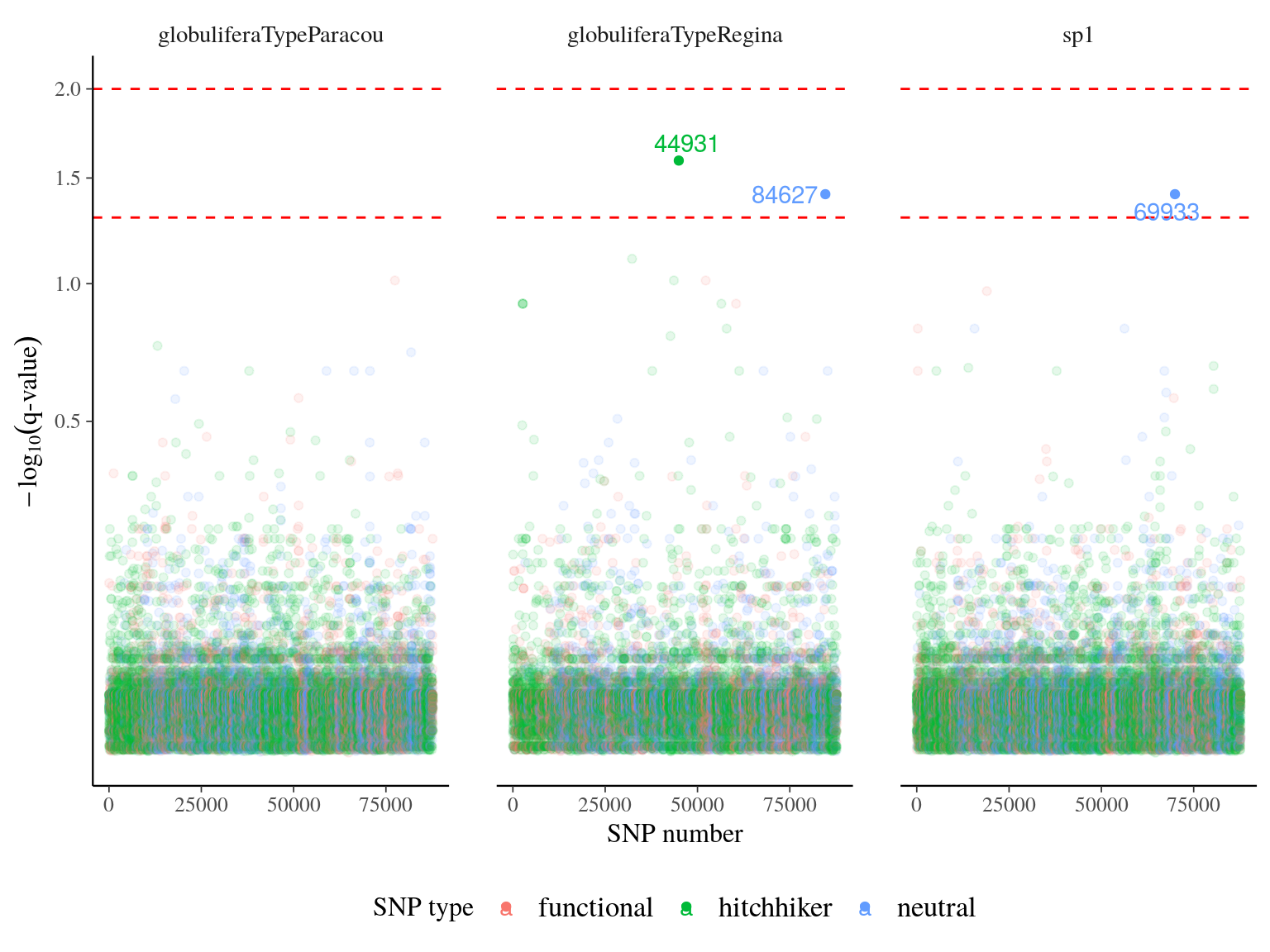
Figure 22.21: q-value of major effect SNPs associated to individual growth parameters. SNP were detected using linear mixed models (LMM) with individual kiniship as random effect. SNP significance was assessed with Wald test. q-value was obtained correcting mutlitple testing with false discovery rate.
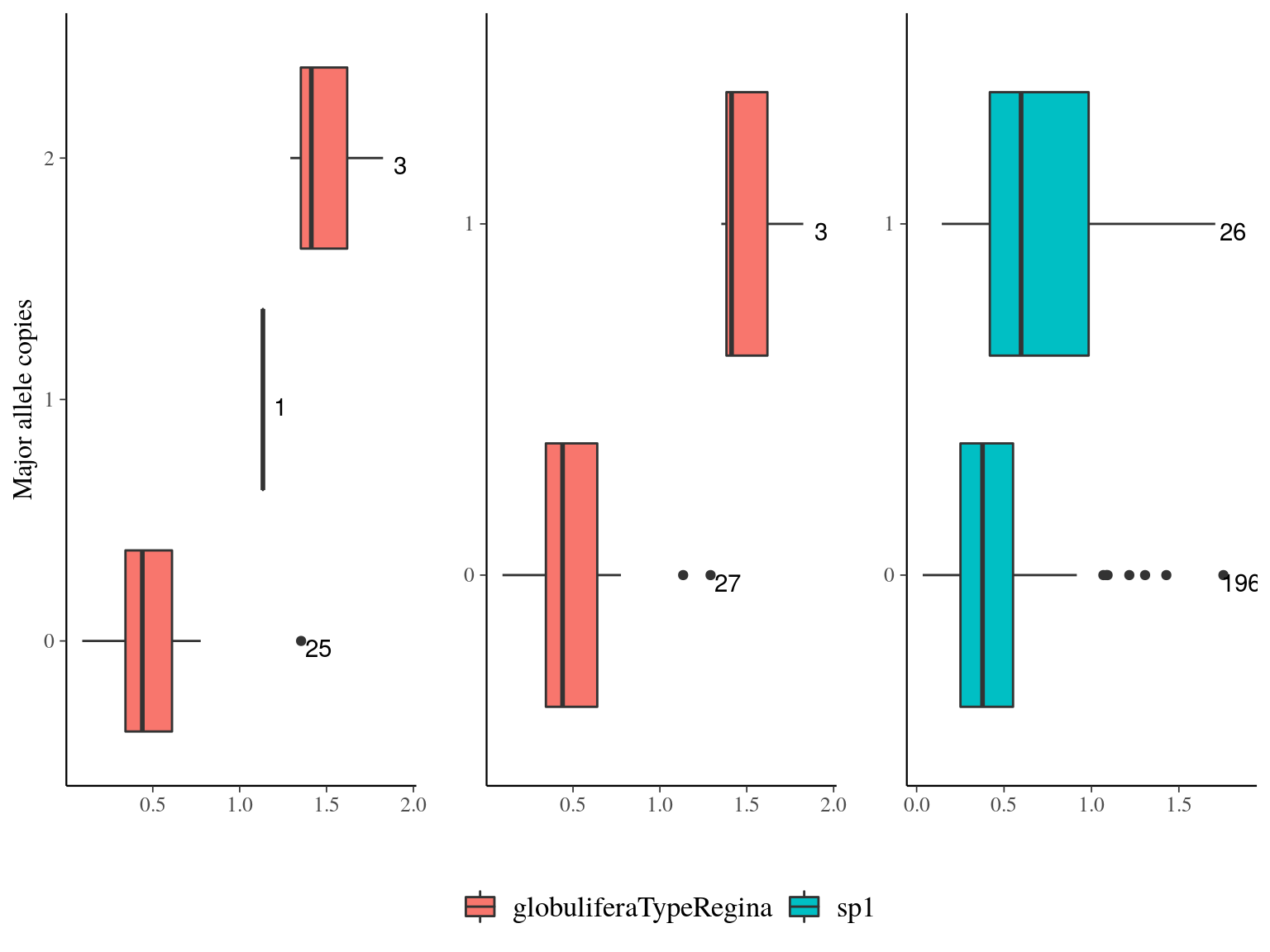
Figure 22.22: Growth parameters distribution for outliers genes.