Chapter 2 Olsson et al. (2017) scaffolds preparation
The African genome scaffolds from Olsson et al. (2017)…
2.1 Renaming
We renamed scaffolds from Olsson using following code : Olsson_2017_[scaffold name].
2.2 Removing scaffolds with multimatch blasted consensus sequence from Torroba-Balmori et al. (unpublished)
We will again use the consensus sequence for French Guianan reads from Torroba-Balmori et al. (unpublished), by blasting them on scaffolds from Olsson et al. (2017) with blastn.
cd ~/Documents/BIOGECO/PhD/data/Symphonia_Genomes/Olsson_2016
cd Olsson2017
makeblastdb -in Olsson2017.fa -parse_seqids -dbtype nucl
cd ..
query=~/Documents/BIOGECO/PhD/data/Symphonia_Torroba/assembly/symphoGbS2_outfiles/symphoGbS2.firstline.fasta
blastn -db Olsson2017/Olsson2017.fa -query $query -out blast_consensus_torroba2.txt -evalue 1e-10 -best_hit_score_edge 0.05 -best_hit_overhang 0.25 -outfmt 6 -perc_identity 75 -max_target_seqs 10blast <- read_tsv(file.path(path, "Olsson_2016", "blast_consensus_torroba2.txt"), col_names = F)
names(blast) <- c("Read", "Scaffold", "Perc_Ident", "Alignment_length", "Mismatches",
"Gap_openings", "R_start", "R_end", "S_start", "S_end", "E", "Bits")
write_file(paste(unique(blast$Scaffold), collapse = "\n"),
file.path(path, "Olsson_2016", "selected_scaffolds_blast_consensus2.list"))We finally have most of scaffolds with on match in a broad range of sizes (from 100 bp to 33.2 kbp). In total 688 scaffolds from Olsson et al. (2017) match consensus sequences from Torroba-Balmori et al. (unpublished). But several scaffolds obtained multiple matches that we cannot use for probes. We will thus exclude the whole scaffold if the scaffold is shorter than 2000 bp or the scaffold region matching the raw read if the scaffold is longer than 2000 bp.
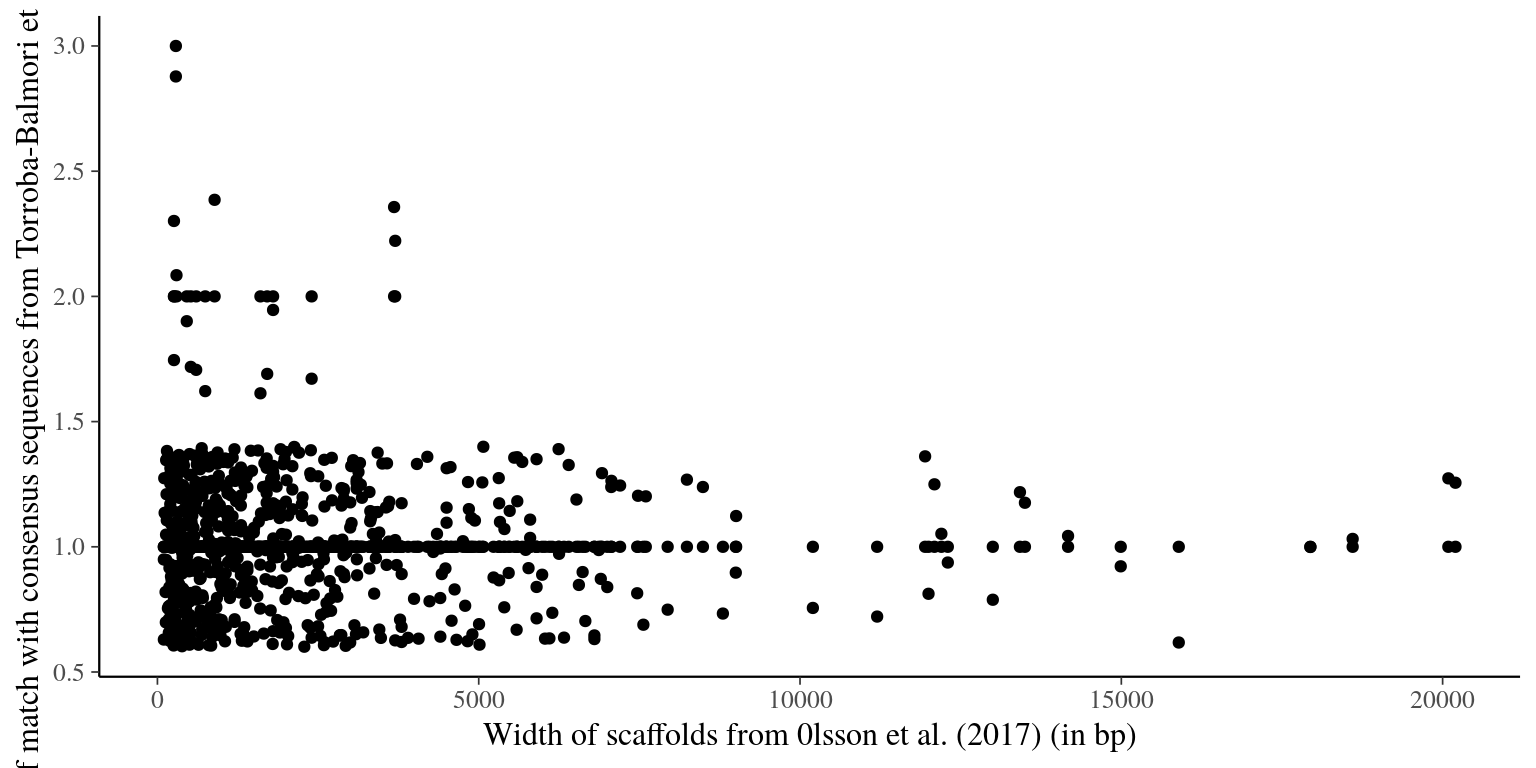
Figure 2.1: Number of match with Torroba consensus reads vs gene width.
| Scaffold | width | remove | cut |
|---|---|---|---|
| Olsson_2017_deg7180004393135 | 2380 | 1700-1631 | |
| Olsson_2017_deg7180004374686 | 2120 | 2073-1992 | |
| Olsson_2017_scf7180005372912 | 3152 | 3102-3152 | |
| Olsson_2017_scf7180005387046 | 2048 | 174-256 | |
| Olsson_2017_scf7180005323991 | 2482 | 2330-2267 |
Following scaffolds will be removed due to multitple matches and a length \(<200bp\): deg7180004378417, deg7180003744575, deg7180002657883, deg7180004705895, deg7180004369764, deg7180002776754, deg7180004453462, deg7180004453461, deg7180002668453, deg7180005298947, deg7180003723902, deg7180005298948, deg7180004372504, deg7180002659849, deg7180004372505, deg7180004377385, deg7180003260802, deg7180003625436, deg7180004705895, deg7180002776754, deg7180004705894, deg7180002852093, deg7180004822905, deg7180005023024, deg7180004478675, deg7180004428004, deg7180004428003, deg7180004507379, deg7180002656221, deg7180004374687, deg7180004372498, deg7180004372497, deg7180002654368, deg7180002674357, deg7180004700334, deg7180004899808, deg7180004899808, deg7180002726303, scf7180005372913, deg7180005163225, deg7180003214542, scf7180005400822, deg7180005163224, deg7180003138164, deg7180004981997, deg7180004981996, deg7180005171251, deg7180005106503, deg7180003910181, deg7180005026532, deg7180003853280, deg7180004724986, deg7180005246885, deg7180004710959, deg7180004681149, deg7180004580422, deg7180004472718, deg7180003290510, deg7180005004768, deg7180004756559, scf7180005435685, deg7180004725719, deg7180004599019, deg7180004599018, deg7180002749392, deg7180002739372, deg7180004754314, deg7180004847375, deg7180004580009, deg7180004386399, deg7180004377195, deg7180004377194, deg7180004399409, deg7180004392029, deg7180004385805, deg7180004386398, deg7180002816623, deg7180002985310, scf7180005421751, deg7180004374725, deg7180004372798, deg7180004374726, deg7180002668107, deg7180003199928, deg7180003093903, deg7180003310549, deg7180004796671, deg7180003505925, deg7180002988969. And other will be cut (see table 2.1).
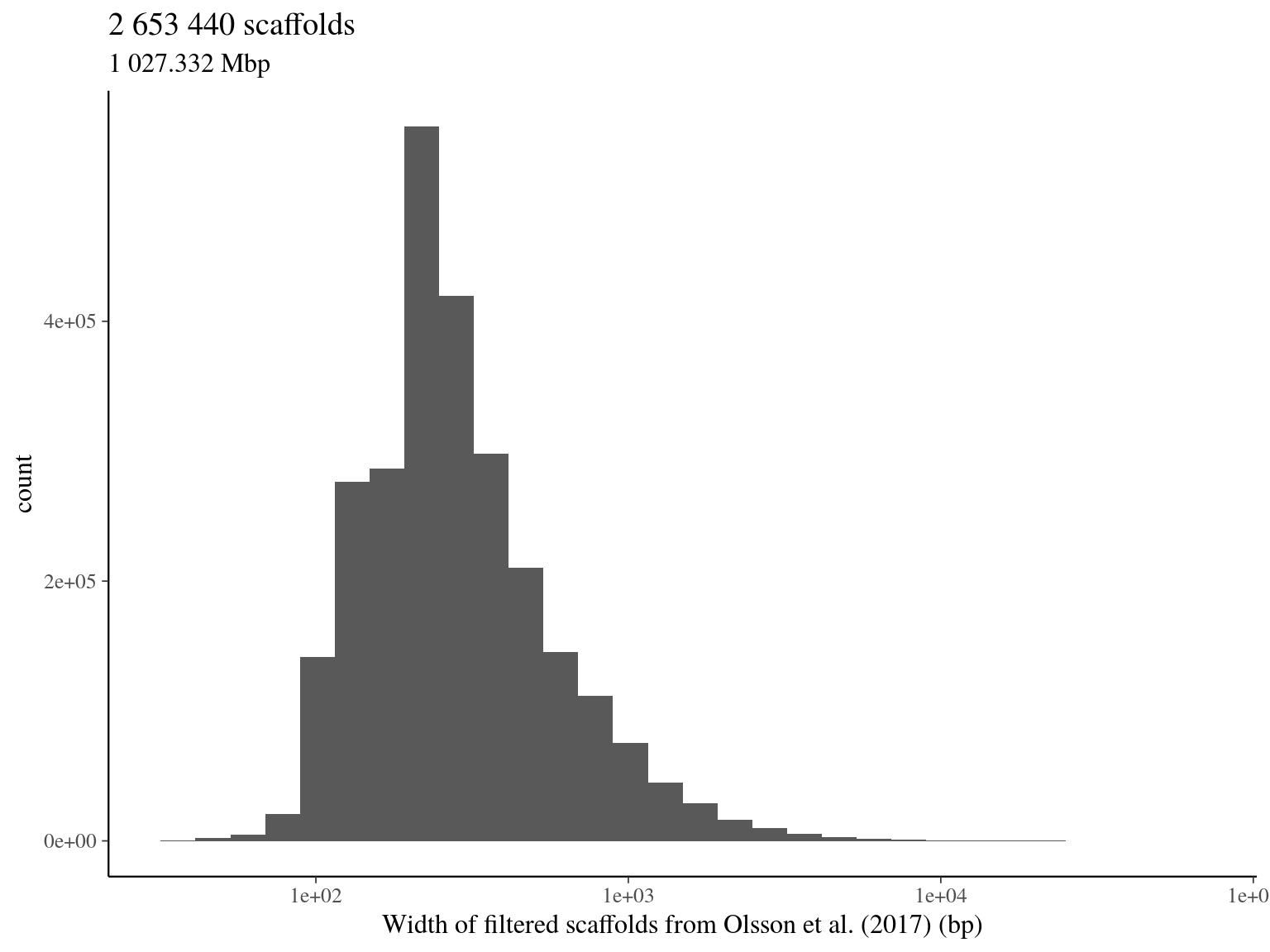
2.3 Total filtered scaffolds
References
Olsson, S., Seoane-Zonjic, P., Bautista, R., Claros, M.G., González-Martínez, S.C., Scotti, I., Scotti-Saintagne, C., Hardy, O.J. & Heuertz, M. (2017). Development of genomic tools in a widespread tropical tree, Symphonia globulifera L.f.: a new low-coverage draft genome, SNP and SSR markers. Molecular Ecology Resources, 17, 614–630. Retrieved from http://doi.wiley.com/10.1111/1755-0998.12605